Mol:FL3FADCS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.7842 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7842 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7842 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7842 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0698 -2.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0698 -2.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3554 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3554 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3554 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3554 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0698 -0.5251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0698 -0.5251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6409 -2.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6409 -2.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0736 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0736 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0736 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0736 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6409 -0.5251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6409 -0.5251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6409 -2.8181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6409 -2.8181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4984 -0.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4984 -0.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8992 2.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8992 2.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8315 1.7630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8315 1.7630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8116 0.8527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8116 0.8527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0833 0.0805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0833 0.0805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3207 0.4360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3207 0.4360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4350 1.2675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4350 1.2675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0144 2.9996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0144 2.9996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5993 2.4956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5993 2.4956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5165 0.3850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5165 0.3850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0698 -2.9996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0698 -2.9996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9467 -0.3927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9467 -0.3927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6995 -0.8274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6995 -0.8274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4523 -0.3927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4523 -0.3927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4523 0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4523 0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6995 0.9112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6995 0.9112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9467 0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9467 0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2047 0.9110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2047 0.9110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0272 0.4790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0272 0.4790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3527 0.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3527 0.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5668 0.8384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5668 0.8384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3527 1.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3527 1.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0272 1.9813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0272 1.9813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8132 1.2324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8132 1.2324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8145 2.5687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8145 2.5687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3841 2.2459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3841 2.2459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4599 1.6411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4599 1.6411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6767 0.4060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6767 0.4060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7474 1.5484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7474 1.5484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0663 0.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0663 0.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5645 -1.0348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5645 -1.0348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6767 -1.6769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6767 -1.6769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 31 21 1 0 0 0 0 | + | 31 21 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 18 40 1 0 0 0 0 | + | 18 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.6876 -0.2809 | + | M SBV 1 45 -0.6876 -0.2809 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 -1.1122 0.6421 | + | M SBV 2 47 -1.1122 0.6421 |
| − | S SKP 5 | + | S SKP 5 |
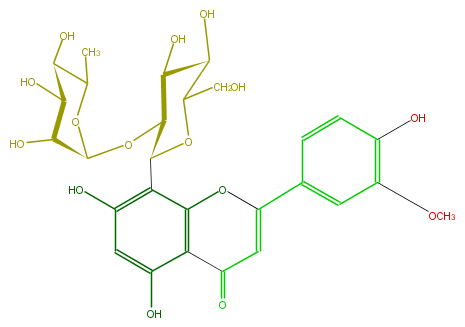
| − | ID FL3FADCS0012 | + | ID FL3FADCS0012 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES OC(C1O)C(OC(O5)C(O)C(O)C(C(C)5)O)C(c(c4O)c(c(c(O)c4)2)OC(c(c3)ccc(O)c(OC)3)=CC2=O)OC(CO)1 | + | SMILES OC(C1O)C(OC(O5)C(O)C(O)C(C(C)5)O)C(c(c4O)c(c(c(O)c4)2)OC(c(c3)ccc(O)c(OC)3)=CC2=O)OC(CO)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.7842 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7842 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0698 -2.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3554 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3554 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0698 -0.5251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6409 -2.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0736 -1.7625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0736 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6409 -0.5251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6409 -2.8181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4984 -0.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8992 2.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8315 1.7630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8116 0.8527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0833 0.0805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3207 0.4360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4350 1.2675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0144 2.9996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5993 2.4956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5165 0.3850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0698 -2.9996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9467 -0.3927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6995 -0.8274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4523 -0.3927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4523 0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6995 0.9112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9467 0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2047 0.9110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0272 0.4790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3527 0.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5668 0.8384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3527 1.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0272 1.9813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8132 1.2324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8145 2.5687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3841 2.2459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4599 1.6411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6767 0.4060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7474 1.5484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0663 0.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5645 -1.0348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6767 -1.6769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
26 29 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
35 38 1 0 0 0 0
30 39 1 0 0 0 0
31 21 1 0 0 0 0
40 41 1 0 0 0 0
18 40 1 0 0 0 0
42 43 1 0 0 0 0
25 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.6876 -0.2809
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 -1.1122 0.6421
S SKP 5
ID FL3FADCS0012
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES OC(C1O)C(OC(O5)C(O)C(O)C(C(C)5)O)C(c(c4O)c(c(c(O)c4)2)OC(c(c3)ccc(O)c(OC)3)=CC2=O)OC(CO)1
M END