Mol:FL3FACGS0054
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.1848 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1848 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1848 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1848 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6358 -0.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6358 -0.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0869 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0869 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0869 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0869 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6358 0.7718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6358 0.7718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5379 -0.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5379 -0.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9890 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9890 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9890 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9890 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5379 0.7718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5379 0.7718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5379 -0.6759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5379 -0.6759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4399 0.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4399 0.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8996 0.5063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8996 0.5063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3594 0.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3594 0.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3594 1.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3594 1.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8996 1.5680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8996 1.5680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4399 1.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4399 1.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3347 0.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3347 0.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6358 -0.7896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6358 -0.7896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9537 1.6457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9537 1.6457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8996 2.0988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8996 2.0988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9330 -0.9927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9330 -0.9927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4173 -1.6733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4173 -1.6733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6748 -1.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6748 -1.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0416 -1.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0416 -1.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4790 -0.8561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4790 -0.8561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2375 -1.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2375 -1.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5095 -1.3255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5095 -1.3255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2211 -1.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2211 -1.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2494 -2.0988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2494 -2.0988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5881 -0.6541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5881 -0.6541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5881 -0.1177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5881 -0.1177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2605 0.2705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2605 0.2705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8179 -0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8179 -0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3733 0.2692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3733 0.2692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9537 -0.0659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9537 -0.0659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3733 0.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3733 0.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2605 0.8546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2605 0.8546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 33 38 2 0 0 0 0 | + | 33 38 2 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACGS0054 | + | ID FL3FACGS0054 |
| − | KNApSAcK_ID C00004314 | + | KNApSAcK_ID C00004314 |
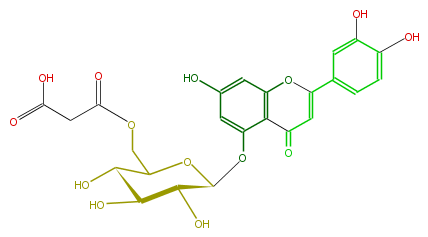
| − | NAME Luteolin 5-(6''-malonylglucoside) | + | NAME Luteolin 5-(6''-malonylglucoside) |
| − | CAS_RN 130733-27-6 | + | CAS_RN 130733-27-6 |
| − | FORMULA C24H22O14 | + | FORMULA C24H22O14 |
| − | EXACTMASS 534.100955412 | + | EXACTMASS 534.100955412 |
| − | AVERAGEMASS 534.42308 | + | AVERAGEMASS 534.42308 |
| − | SMILES O=C(CC(O)=O)OCC(C(O)4)OC(C(C(O)4)O)Oc(c12)cc(cc1OC(c(c3)cc(c(c3)O)O)=CC2=O)O | + | SMILES O=C(CC(O)=O)OCC(C(O)4)OC(C(C(O)4)O)Oc(c12)cc(cc1OC(c(c3)cc(c(c3)O)O)=CC2=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
0.1848 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1848 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6358 -0.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0869 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0869 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6358 0.7718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5379 -0.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9890 -0.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9890 0.5114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5379 0.7718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5379 -0.6759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4399 0.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8996 0.5063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3594 0.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3594 1.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8996 1.5680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4399 1.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3347 0.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6358 -0.7896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9537 1.6457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8996 2.0988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9330 -0.9927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4173 -1.6733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6748 -1.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0416 -1.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4790 -0.8561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2375 -1.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5095 -1.3255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2211 -1.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2494 -2.0988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5881 -0.6541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5881 -0.1177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2605 0.2705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8179 -0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3733 0.2692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9537 -0.0659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3733 0.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2605 0.8546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
35 37 1 0 0 0 0
33 38 2 0 0 0 0
25 19 1 0 0 0 0
S SKP 8
ID FL3FACGS0054
KNApSAcK_ID C00004314
NAME Luteolin 5-(6''-malonylglucoside)
CAS_RN 130733-27-6
FORMULA C24H22O14
EXACTMASS 534.100955412
AVERAGEMASS 534.42308
SMILES O=C(CC(O)=O)OCC(C(O)4)OC(C(C(O)4)O)Oc(c12)cc(cc1OC(c(c3)cc(c(c3)O)O)=CC2=O)O
M END