Mol:FL3FACGS0033
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6561 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6561 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6561 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6561 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0357 -2.4267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0357 -2.4267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5846 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5846 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5846 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5846 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0357 -0.9940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0357 -0.9940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2050 -2.4267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2050 -2.4267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8254 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8254 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8254 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8254 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2050 -0.9940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2050 -0.9940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2050 -3.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2050 -3.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4455 -0.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4455 -0.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0778 -1.3592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0778 -1.3592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7100 -0.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7100 -0.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7100 -0.2640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7100 -0.2640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0778 0.1010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0778 0.1010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4455 -0.2640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4455 -0.2640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3706 -0.9397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3706 -0.9397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0357 -3.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0357 -3.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5274 0.2079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5274 0.2079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0335 -0.9218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0335 -0.9218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5370 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5370 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8221 -1.2990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8221 -1.2990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1324 -1.2916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1324 -1.2916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6336 -0.7903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6336 -0.7903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3639 -1.0524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3639 -1.0524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0226 -0.5013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0226 -0.5013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3108 -1.6147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3108 -1.6147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0972 -1.7680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0972 -1.7680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0778 0.8309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0778 0.8309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1894 2.5800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1894 2.5800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0770 1.7656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0770 1.7656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7791 1.4567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7791 1.4567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2721 0.9743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2721 0.9743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2722 1.6832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2722 1.6832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5704 2.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5704 2.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6104 3.0189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6104 3.0189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6490 2.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6490 2.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9345 0.6177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9345 0.6177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6025 2.5821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6025 2.5821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0920 3.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0920 3.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5046 2.9184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5046 2.9184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7145 -0.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7145 -0.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5274 0.0440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5274 0.0440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1860 0.5364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1860 0.5364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 16 30 1 0 0 0 0 | + | 16 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 40 41 42 | + | M SAL 1 3 40 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 46 -0.0321 -0.5680 | + | M SBV 1 46 -0.0321 -0.5680 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 43 44 45 | + | M SAL 2 3 43 44 45 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 47 0.3507 -0.9553 | + | M SBV 2 47 0.3507 -0.9553 |
| − | S SKP 5 | + | S SKP 5 |
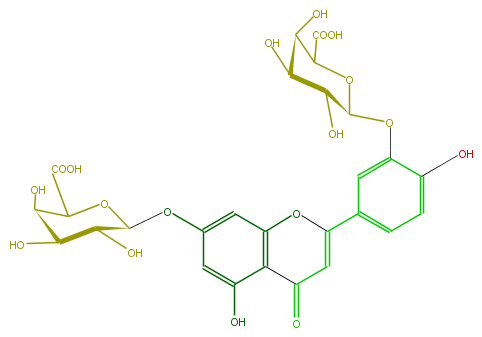
| − | ID FL3FACGS0033 | + | ID FL3FACGS0033 |
| − | FORMULA C27H26O18 | + | FORMULA C27H26O18 |
| − | EXACTMASS 638.111914028 | + | EXACTMASS 638.111914028 |
| − | AVERAGEMASS 638.4845399999999 | + | AVERAGEMASS 638.4845399999999 |
| − | SMILES O(C(c(c4)cc(OC(C5O)OC(C(O)C5O)C(O)=O)c(c4)O)=1)c(c2)c(c(cc2OC(O3)C(C(C(O)C3C(O)=O)O)O)O)C(=O)C1 | + | SMILES O(C(c(c4)cc(OC(C5O)OC(C(O)C5O)C(O)=O)c(c4)O)=1)c(c2)c(c(cc2OC(O3)C(C(C(O)C3C(O)=O)O)O)O)C(=O)C1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-0.6561 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6561 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0357 -2.4267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5846 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5846 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0357 -0.9940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2050 -2.4267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8254 -2.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8254 -1.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2050 -0.9940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2050 -3.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4455 -0.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0778 -1.3592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7100 -0.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7100 -0.2640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0778 0.1010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4455 -0.2640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3706 -0.9397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0357 -3.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5274 0.2079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0335 -0.9218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5370 -1.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8221 -1.2990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1324 -1.2916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6336 -0.7903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3639 -1.0524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0226 -0.5013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3108 -1.6147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0972 -1.7680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0778 0.8309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1894 2.5800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0770 1.7656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7791 1.4567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2721 0.9743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2722 1.6832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5704 2.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6104 3.0189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6490 2.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9345 0.6177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6025 2.5821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0920 3.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5046 2.9184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7145 -0.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5274 0.0440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1860 0.5364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
16 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 30 1 0 0 0 0
40 41 2 0 0 0 0
40 42 1 0 0 0 0
36 40 1 0 0 0 0
26 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 40 41 42
M SBL 1 1 46
M SMT 1 COOH
M SBV 1 46 -0.0321 -0.5680
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 43 44 45
M SBL 2 1 47
M SMT 2 ^ COOH
M SBV 2 47 0.3507 -0.9553
S SKP 5
ID FL3FACGS0033
FORMULA C27H26O18
EXACTMASS 638.111914028
AVERAGEMASS 638.4845399999999
SMILES O(C(c(c4)cc(OC(C5O)OC(C(O)C5O)C(O)=O)c(c4)O)=1)c(c2)c(c(cc2OC(O3)C(C(C(O)C3C(O)=O)O)O)O)C(=O)C1
M END