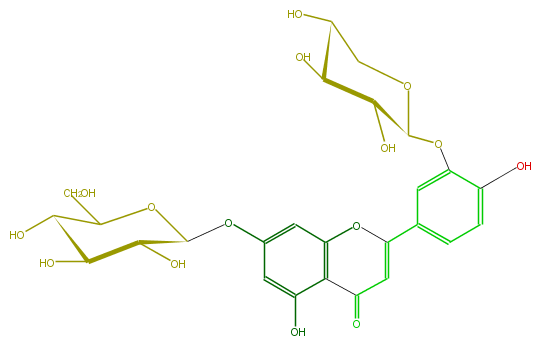
Mol:FL3FACGS0029
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.1313 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1313 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1313 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1313 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4841 -2.4794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4841 -2.4794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0996 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0996 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0996 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0996 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4841 -1.0581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4841 -1.0581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7151 -2.4794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7151 -2.4794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3305 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3305 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3305 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3305 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7151 -1.0581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7151 -1.0581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7151 -3.0336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7151 -3.0336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9458 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9458 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5730 -1.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5730 -1.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2004 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2004 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2004 -0.3339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2004 -0.3339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5730 0.0284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5730 0.0284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9458 -0.3339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9458 -0.3339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8401 -1.0042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8401 -1.0042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4841 -3.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4841 -3.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0113 0.1344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0113 0.1344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3662 -0.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3662 -0.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6626 -1.8022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6626 -1.8022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6495 -1.4083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6495 -1.4083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6719 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6719 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3822 -0.6872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3822 -0.6872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4172 -1.0587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4172 -1.0587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0113 -1.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0113 -1.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4166 -1.8022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4166 -1.8022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9211 -1.8288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9211 -1.8288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3633 0.5674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3633 0.5674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2137 3.0115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2137 3.0115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0546 1.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0546 1.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0496 1.4196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0496 1.4196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7482 0.7358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7482 0.7358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7483 1.7405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7483 1.7405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7538 2.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7538 2.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5530 3.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5530 3.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5852 2.3269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5852 2.3269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2949 0.5038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2949 0.5038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0786 -0.3973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0786 -0.3973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1038 -0.9602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1038 -0.9602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 16 30 1 0 0 0 0 | + | 16 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 0.6614 -0.6614 | + | M SBV 1 45 0.6614 -0.6614 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACGS0029 | + | ID FL3FACGS0029 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES c(c2)(C(=C5)Oc(c3C(=O)5)cc(OC(C(O)4)OC(CO)C(O)C4O)cc3O)cc(c(O)c2)OC(O1)C(O)C(O)C(O)C1 | + | SMILES c(c2)(C(=C5)Oc(c3C(=O)5)cc(OC(C(O)4)OC(CO)C(O)C4O)cc3O)cc(c(O)c2)OC(O1)C(O)C(O)C(O)C1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.1313 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1313 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4841 -2.4794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0996 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0996 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4841 -1.0581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7151 -2.4794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3305 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3305 -1.4134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7151 -1.0581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7151 -3.0336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9458 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5730 -1.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2004 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2004 -0.3339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5730 0.0284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9458 -0.3339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8401 -1.0042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4841 -3.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0113 0.1344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3662 -0.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6626 -1.8022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6495 -1.4083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6719 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3822 -0.6872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4172 -1.0587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0113 -1.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4166 -1.8022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9211 -1.8288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3633 0.5674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2137 3.0115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0546 1.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0496 1.4196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7482 0.7358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7483 1.7405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7538 2.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5530 3.1886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5852 2.3269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2949 0.5038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0786 -0.3973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1038 -0.9602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
16 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 30 1 0 0 0 0
40 41 1 0 0 0 0
26 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 0.6614 -0.6614
S SKP 5
ID FL3FACGS0029
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES c(c2)(C(=C5)Oc(c3C(=O)5)cc(OC(C(O)4)OC(CO)C(O)C4O)cc3O)cc(c(O)c2)OC(O1)C(O)C(O)C(O)C1
M END