Mol:FL3FACGS0022
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5406 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5406 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5406 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5406 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0901 -1.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0901 -1.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7210 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7210 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7210 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7210 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0901 0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0901 0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3519 -1.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3519 -1.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9826 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9826 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9826 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9826 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3519 0.2217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3519 0.2217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3459 -1.9986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3459 -1.9986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6133 0.2215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6133 0.2215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2563 -0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2563 -0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8992 0.2215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8992 0.2215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8992 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8992 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2563 1.3350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2563 1.3350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6133 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6133 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2672 0.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2672 0.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0901 -1.9622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0901 -1.9622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7304 1.4439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7304 1.4439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2563 2.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2563 2.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0010 0.2917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0010 0.2917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3945 -0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3945 -0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6676 -0.0905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6676 -0.0905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9661 -0.0829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9661 -0.0829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4759 0.4268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4759 0.4268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2184 0.1603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2184 0.1603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7304 -0.0135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7304 -0.0135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1893 -0.3372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1893 -0.3372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9945 -0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9945 -0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9235 -1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9235 -1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3615 -1.6802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3615 -1.6802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4531 -1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4531 -1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0032 -1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0032 -1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9905 -0.8530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9905 -0.8530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3482 -2.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3482 -2.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4511 -2.1646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4511 -2.1646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7046 0.6439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7046 0.6439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7712 1.0216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7712 1.0216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3508 -1.2712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3508 -1.2712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6258 -0.8775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6258 -0.8775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 18 1 0 0 0 0 | + | 25 18 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 39 38 1 0 0 0 0 | + | 39 38 1 0 0 0 0 |
| − | 27 38 1 0 0 0 0 | + | 27 38 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 43 0.4862 -0.4837 | + | M SBV 1 43 0.4862 -0.4837 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 44 -0.0107 -0.4090 | + | M SBV 2 44 -0.0107 -0.4090 |
| − | S SKP 5 | + | S SKP 5 |
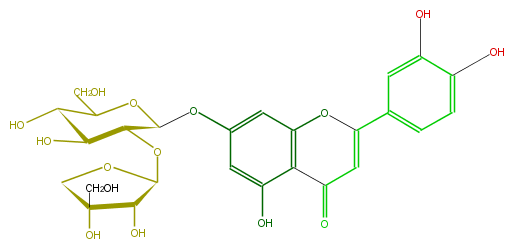
| − | ID FL3FACGS0022 | + | ID FL3FACGS0022 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES c(C(=O)5)(c1OC(=C5)c(c4)cc(c(c4)O)O)c(O)cc(OC(O2)C(OC(O3)C(O)C(O)(C3)CO)C(O)C(C(CO)2)O)c1 | + | SMILES c(C(=O)5)(c1OC(=C5)c(c4)cc(c(c4)O)O)c(O)cc(OC(O2)C(OC(O3)C(O)C(O)(C3)CO)C(O)C(C(CO)2)O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.5406 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5406 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0901 -1.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7210 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7210 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0901 0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3519 -1.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9826 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9826 -0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3519 0.2217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3459 -1.9986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6133 0.2215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2563 -0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8992 0.2215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8992 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2563 1.3350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6133 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2672 0.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0901 -1.9622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7304 1.4439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2563 2.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0010 0.2917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3945 -0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6676 -0.0905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9661 -0.0829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4759 0.4268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2184 0.1603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7304 -0.0135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1893 -0.3372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9945 -0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9235 -1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3615 -1.6802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4531 -1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0032 -1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9905 -0.8530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3482 -2.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4511 -2.1646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7046 0.6439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7712 1.0216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3508 -1.2712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6258 -0.8775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 18 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 31 1 0 0 0 0
32 36 1 0 0 0 0
33 37 1 0 0 0 0
34 30 1 0 0 0 0
39 38 1 0 0 0 0
27 38 1 0 0 0 0
32 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 ^ CH2OH
M SBV 1 43 0.4862 -0.4837
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 CH2OH
M SBV 2 44 -0.0107 -0.4090
S SKP 5
ID FL3FACGS0022
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES c(C(=O)5)(c1OC(=C5)c(c4)cc(c(c4)O)O)c(O)cc(OC(O2)C(OC(O3)C(O)C(O)(C3)CO)C(O)C(C(CO)2)O)c1
M END