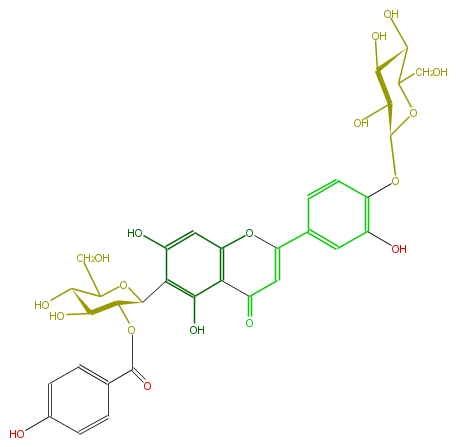
Mol:FL3FACDS0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 57 0 0 0 0 0 0 0 0999 V2000 | + | 52 57 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4356 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4356 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4356 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4356 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8793 -1.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8793 -1.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3230 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3230 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3230 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3230 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8793 -0.1817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8793 -0.1817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2333 -1.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2333 -1.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7896 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7896 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7896 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7896 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2333 -0.1817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2333 -0.1817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2333 -1.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2333 -1.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9917 -0.1818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9917 -0.1818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8793 -2.1086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8793 -2.1086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4075 -0.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4075 -0.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9938 -0.4997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9938 -0.4997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5800 -0.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5800 -0.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5800 0.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5800 0.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9938 0.8541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9938 0.8541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4075 0.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4075 0.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1658 0.8539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1658 0.8539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3268 -1.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3268 -1.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9557 -1.8002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9557 -1.8002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4212 -1.5924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4212 -1.5924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9054 -1.5868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9054 -1.5868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2802 -1.2119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2802 -1.2119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7478 -1.4587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7478 -1.4587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8407 -1.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8407 -1.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5343 -1.8284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5343 -1.8284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1149 -2.1065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1149 -2.1065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3879 3.5134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3879 3.5134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7824 3.0546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7824 3.0546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0393 2.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0393 2.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0462 1.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0462 1.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5095 2.2197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5095 2.2197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2044 2.7977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2044 2.7977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9894 4.2037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9894 4.2037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7475 3.7698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7475 3.7698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4038 2.0154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4038 2.0154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1149 -2.9172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1149 -2.9172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7950 -3.2371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7950 -3.2371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6012 -3.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6012 -3.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1819 -2.8626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1819 -2.8626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7626 -3.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7626 -3.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7626 -3.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7626 -3.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1819 -4.2037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1819 -4.2037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6012 -3.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6012 -3.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3425 -4.2033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3425 -4.2033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1658 -0.4995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1658 -0.4995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1463 -0.6746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1463 -0.6746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3494 -0.8881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3494 -0.8881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6281 3.0423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6281 3.0423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3425 2.6298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3425 2.6298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 14 9 1 0 0 0 0 | + | 14 9 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 29 39 1 0 0 0 0 | + | 29 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 44 47 1 0 0 0 0 | + | 44 47 1 0 0 0 0 |
| − | 16 48 1 0 0 0 0 | + | 16 48 1 0 0 0 0 |
| − | 26 49 1 0 0 0 0 | + | 26 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 35 51 1 0 0 0 0 | + | 35 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 49 50 | + | M SAL 1 2 49 50 |
| − | M SBL 1 1 54 | + | M SBL 1 1 54 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 54 -7.4812 6.5925 | + | M SBV 1 54 -7.4812 6.5925 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 51 52 | + | M SAL 2 2 51 52 |
| − | M SBL 2 1 56 | + | M SBL 2 1 56 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 56 -6.6591 6.0529 | + | M SBV 2 56 -6.6591 6.0529 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACDS0016 | + | ID FL3FACDS0016 |
| − | KNApSAcK_ID C00006323 | + | KNApSAcK_ID C00006323 |
| − | NAME Isorientin 4'-O-glucoside 2''-O-p-hydroxybenzoagte | + | NAME Isorientin 4'-O-glucoside 2''-O-p-hydroxybenzoagte |
| − | CAS_RN 67308-39-8 | + | CAS_RN 67308-39-8 |
| − | FORMULA C34H34O18 | + | FORMULA C34H34O18 |
| − | EXACTMASS 730.174514284 | + | EXACTMASS 730.174514284 |
| − | AVERAGEMASS 730.6229599999999 | + | AVERAGEMASS 730.6229599999999 |
| − | SMILES OC(C6O)C(OC(C6O)Oc(c(O)1)ccc(C(O5)=CC(c(c25)c(O)c(C(C3OC(c(c4)ccc(O)c4)=O)OC(C(O)C3O)CO)c(c2)O)=O)c1)CO | + | SMILES OC(C6O)C(OC(C6O)Oc(c(O)1)ccc(C(O5)=CC(c(c25)c(O)c(C(C3OC(c(c4)ccc(O)c4)=O)OC(C(O)C3O)CO)c(c2)O)=O)c1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 57 0 0 0 0 0 0 0 0999 V2000
-1.4356 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4356 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8793 -1.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3230 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3230 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8793 -0.1817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2333 -1.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7896 -1.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7896 -0.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2333 -0.1817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2333 -1.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9917 -0.1818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8793 -2.1086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4075 -0.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9938 -0.4997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5800 -0.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5800 0.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9938 0.8541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4075 0.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1658 0.8539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3268 -1.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9557 -1.8002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4212 -1.5924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9054 -1.5868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2802 -1.2119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7478 -1.4587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8407 -1.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5343 -1.8284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1149 -2.1065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3879 3.5134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7824 3.0546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0393 2.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0462 1.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5095 2.2197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2044 2.7977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9894 4.2037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7475 3.7698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4038 2.0154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1149 -2.9172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7950 -3.2371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6012 -3.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1819 -2.8626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7626 -3.1979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7626 -3.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1819 -4.2037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6012 -3.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3425 -4.2033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1658 -0.4995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1463 -0.6746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3494 -0.8881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6281 3.0423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3425 2.6298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
14 9 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 2 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 20 1 0 0 0 0
29 39 1 0 0 0 0
39 40 2 0 0 0 0
39 41 1 0 0 0 0
41 42 2 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 41 1 0 0 0 0
44 47 1 0 0 0 0
16 48 1 0 0 0 0
26 49 1 0 0 0 0
49 50 1 0 0 0 0
35 51 1 0 0 0 0
51 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 49 50
M SBL 1 1 54
M SMT 1 ^CH2OH
M SBV 1 54 -7.4812 6.5925
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 51 52
M SBL 2 1 56
M SMT 2 CH2OH
M SBV 2 56 -6.6591 6.0529
S SKP 8
ID FL3FACDS0016
KNApSAcK_ID C00006323
NAME Isorientin 4'-O-glucoside 2''-O-p-hydroxybenzoagte
CAS_RN 67308-39-8
FORMULA C34H34O18
EXACTMASS 730.174514284
AVERAGEMASS 730.6229599999999
SMILES OC(C6O)C(OC(C6O)Oc(c(O)1)ccc(C(O5)=CC(c(c25)c(O)c(C(C3OC(c(c4)ccc(O)c4)=O)OC(C(O)C3O)CO)c(c2)O)=O)c1)CO
M END