Mol:FL3FACDS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.5844 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5844 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5844 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5844 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1407 -1.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1407 -1.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6970 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6970 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6970 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6970 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1407 -0.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1407 -0.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2533 -1.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2533 -1.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8096 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8096 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8096 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8096 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2533 -0.5248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2533 -0.5248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2533 -2.3103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2533 -2.3103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1407 -2.4516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1407 -2.4516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4895 -0.4218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4895 -0.4218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0757 -0.7602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0757 -0.7602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6619 -0.4218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6619 -0.4218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6619 0.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6619 0.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0757 0.5936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0757 0.5936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4895 0.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4895 0.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3270 0.6391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3270 0.6391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0121 -0.5155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0121 -0.5155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6817 1.7528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6817 1.7528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1973 1.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1973 1.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7129 1.4847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7129 1.4847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4142 1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4142 1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9192 1.8559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9192 1.8559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3623 1.5878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3623 1.5878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1699 1.4573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1699 1.4573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5550 1.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5550 1.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0223 0.9488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0223 0.9488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0757 1.6919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0757 1.6919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0308 -1.7535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0308 -1.7535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5152 -2.3310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5152 -2.3310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9996 -2.0216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9996 -2.0216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2983 -2.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2983 -2.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7933 -1.6503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7933 -1.6503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3502 -1.9185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3502 -1.9185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5426 -2.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5426 -2.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1575 -2.4135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1575 -2.4135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6902 -2.5574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6902 -2.5574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9403 -1.3284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9403 -1.3284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5895 -0.8212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5895 -0.8212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8152 -1.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8152 -1.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2996 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2996 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7839 -1.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7839 -1.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0827 -1.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0827 -1.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5777 -1.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5777 -1.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1346 -1.2791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1346 -1.2791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3270 -1.4096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3270 -1.4096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9419 -1.7741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9419 -1.7741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4746 -1.9181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4746 -1.9181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0093 1.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0093 1.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4260 2.5574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4260 2.5574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4876 -0.8928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4876 -0.8928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0709 -0.3094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0709 -0.3094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 17 30 1 0 0 0 0 | + | 17 30 1 0 0 0 0 |
| − | 30 24 1 0 0 0 0 | + | 30 24 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 41 40 1 0 0 0 0 | + | 41 40 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 41 1 0 0 0 0 | + | 45 41 1 0 0 0 0 |
| − | 34 2 1 0 0 0 0 | + | 34 2 1 0 0 0 0 |
| − | 26 51 1 0 0 0 0 | + | 26 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 47 53 1 0 0 0 0 | + | 47 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 56 -7.4988 4.6759 | + | M SBV 1 56 -7.4988 4.6759 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 58 -7.4988 4.6759 | + | M SBV 2 58 -7.4988 4.6759 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACDS0012 | + | ID FL3FACDS0012 |
| − | KNApSAcK_ID C00006314 | + | KNApSAcK_ID C00006314 |
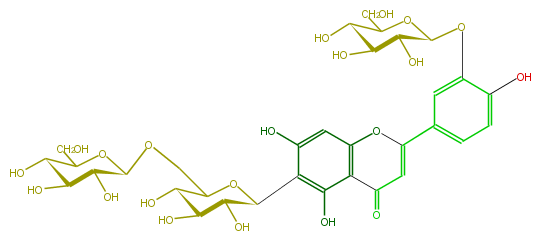
| − | NAME Isoorientin 3',6''-di-O-glucoside | + | NAME Isoorientin 3',6''-di-O-glucoside |
| − | CAS_RN 98568-81-1 | + | CAS_RN 98568-81-1 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES O(C(c(c6O)c(c(c(c6)5)C(=O)C=C(O5)c(c4)cc(c(c4)O)OC(O3)C(C(O)C(O)C(CO)3)O)O)1)C(COC(C2O)OC(C(C2O)O)CO)C(O)C(C(O)1)O | + | SMILES O(C(c(c6O)c(c(c(c6)5)C(=O)C=C(O5)c(c4)cc(c(c4)O)OC(O3)C(C(O)C(O)C(CO)3)O)O)1)C(COC(C2O)OC(C(C2O)O)CO)C(O)C(C(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
0.5844 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5844 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1407 -1.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6970 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6970 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1407 -0.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2533 -1.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8096 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8096 -0.8460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2533 -0.5248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2533 -2.3103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1407 -2.4516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4895 -0.4218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0757 -0.7602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6619 -0.4218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6619 0.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0757 0.5936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4895 0.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3270 0.6391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0121 -0.5155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6817 1.7528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1973 1.1753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7129 1.4847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4142 1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9192 1.8559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3623 1.5878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1699 1.4573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5550 1.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0223 0.9488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0757 1.6919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0308 -1.7535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5152 -2.3310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9996 -2.0216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2983 -2.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7933 -1.6503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3502 -1.9185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5426 -2.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1575 -2.4135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6902 -2.5574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9403 -1.3284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5895 -0.8212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8152 -1.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2996 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7839 -1.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0827 -1.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5777 -1.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1346 -1.2791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3270 -1.4096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9419 -1.7741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4746 -1.9181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0093 1.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4260 2.5574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4876 -0.8928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0709 -0.3094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
17 30 1 0 0 0 0
30 24 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
41 40 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 41 1 0 0 0 0
34 2 1 0 0 0 0
26 51 1 0 0 0 0
51 52 1 0 0 0 0
47 53 1 0 0 0 0
53 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 56
M SMT 1 ^CH2OH
M SBV 1 56 -7.4988 4.6759
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 58
M SMT 2 ^CH2OH
M SBV 2 58 -7.4988 4.6759
S SKP 8
ID FL3FACDS0012
KNApSAcK_ID C00006314
NAME Isoorientin 3',6''-di-O-glucoside
CAS_RN 98568-81-1
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES O(C(c(c6O)c(c(c(c6)5)C(=O)C=C(O5)c(c4)cc(c(c4)O)OC(O3)C(C(O)C(O)C(CO)3)O)O)1)C(COC(C2O)OC(C(C2O)O)CO)C(O)C(C(O)1)O
M END