Mol:FL3FACDS0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7647 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7647 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7647 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7647 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2084 -0.7966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2084 -0.7966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3479 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3479 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3479 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3479 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2084 0.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2084 0.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9042 -0.7966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9042 -0.7966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4605 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4605 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4605 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4605 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9042 0.4881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9042 0.4881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9042 -1.2975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9042 -1.2975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2084 -1.4388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2084 -1.4388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1403 0.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1403 0.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7265 0.2526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7265 0.2526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3127 0.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3127 0.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3127 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3127 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7265 1.6064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7265 1.6064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1403 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1403 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8986 1.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8986 1.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5031 0.7650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5031 0.7650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9262 -0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9262 -0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4106 -1.3800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4106 -1.3800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8950 -1.0706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8950 -1.0706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1937 -1.1531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1937 -1.1531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6887 -0.6993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6887 -0.6993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2456 -0.9675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2456 -0.9675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2940 -1.0148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2940 -1.0148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0530 -1.4625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0530 -1.4625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5856 -1.6064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5856 -1.6064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8986 0.2528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8986 0.2528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7719 0.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7719 0.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2562 0.1257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2562 0.1257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7406 0.4350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7406 0.4350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0394 0.3525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0394 0.3525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5344 0.8063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5344 0.8063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0912 0.5382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0912 0.5382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6042 1.3287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6042 1.3287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8986 0.0432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8986 0.0432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4312 -0.1008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4312 -0.1008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3962 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3962 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1931 -0.3555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1931 -0.3555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7775 1.1318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7775 1.1318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0630 0.7193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0630 0.7193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 15 30 1 0 0 0 0 | + | 15 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 44 -5.9643 7.1654 | + | M SBV 1 44 -5.9643 7.1654 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -5.5000 7.3605 | + | M SBV 2 46 -5.5000 7.3605 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACDS0008 | + | ID FL3FACDS0008 |
| − | KNApSAcK_ID C00006213 | + | KNApSAcK_ID C00006213 |
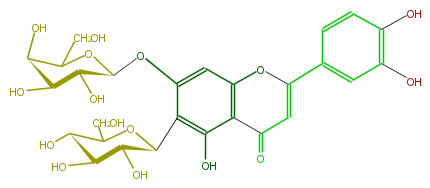
| − | NAME Isoorientin 7-O-galactoside | + | NAME Isoorientin 7-O-galactoside |
| − | CAS_RN 119976-99-7 | + | CAS_RN 119976-99-7 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(O)c3O)OC(C2O)OC(C(O)C2O)CO)(C(O)1)OC(C(O)C(O)1)CO | + | SMILES C(c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(O)c3O)OC(C2O)OC(C(O)C2O)CO)(C(O)1)OC(C(O)C(O)1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.7647 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7647 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2084 -0.7966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3479 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3479 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2084 0.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9042 -0.7966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4605 -0.4755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4605 0.1669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9042 0.4881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9042 -1.2975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2084 -1.4388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1403 0.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7265 0.2526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3127 0.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3127 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7265 1.6064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1403 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8986 1.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5031 0.7650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9262 -0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4106 -1.3800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8950 -1.0706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1937 -1.1531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6887 -0.6993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2456 -0.9675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2940 -1.0148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0530 -1.4625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5856 -1.6064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8986 0.2528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7719 0.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2562 0.1257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7406 0.4350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0394 0.3525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5344 0.8063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0912 0.5382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6042 1.3287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8986 0.0432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4312 -0.1008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3962 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1931 -0.3555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7775 1.1318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0630 0.7193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 2 1 0 0 0 0
15 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 20 1 0 0 0 0
26 40 1 0 0 0 0
40 41 1 0 0 0 0
36 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 ^CH2OH
M SBV 1 44 -5.9643 7.1654
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -5.5000 7.3605
S SKP 8
ID FL3FACDS0008
KNApSAcK_ID C00006213
NAME Isoorientin 7-O-galactoside
CAS_RN 119976-99-7
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(O)c3O)OC(C2O)OC(C(O)C2O)CO)(C(O)1)OC(C(O)C(O)1)CO
M END