Mol:FL3FACCS0030
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.6849 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6849 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6849 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6849 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2412 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2412 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7975 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7975 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7975 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7975 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2412 0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2412 0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3538 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3538 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9101 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9101 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9101 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9101 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3538 0.3725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3538 0.3725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3538 -1.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3538 -1.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2412 -1.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2412 -1.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5900 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5900 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1762 0.1371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1762 0.1371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7624 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7624 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7624 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7624 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1762 1.4909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1762 1.4909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5900 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5900 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4740 1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4740 1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8685 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8685 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3529 -1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3529 -1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8372 -1.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8372 -1.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1360 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1360 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6310 -0.7324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6310 -0.7324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1879 -1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1879 -1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3803 -1.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3803 -1.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9952 -1.4955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9952 -1.4955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1126 0.3818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1126 0.3818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5279 -1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5279 -1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7779 -0.4104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7779 -0.4104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9622 0.1132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9622 0.1132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4466 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4466 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9310 -0.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9310 -0.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2297 -0.2374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2297 -0.2374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7247 0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7247 0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2816 -0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2816 -0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4740 -0.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4740 -0.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0890 -0.5468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0890 -0.5468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6216 -0.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6216 -0.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6636 0.3287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6636 0.3287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3482 0.1373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3482 0.1373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6346 0.3345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6346 0.3345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2180 0.9179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2180 0.9179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 15 41 1 0 0 0 0 | + | 15 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 46 -7.2746 5.6558 | + | M SBV 1 46 -7.2746 5.6558 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACCS0030 | + | ID FL3FACCS0030 |
| − | KNApSAcK_ID C00006211 | + | KNApSAcK_ID C00006211 |
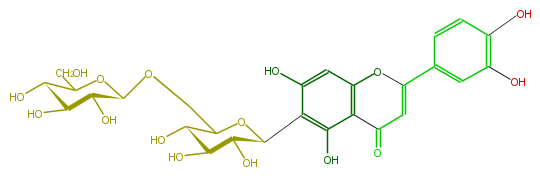
| − | NAME Isoorientin 6''-O-glucoside | + | NAME Isoorientin 6''-O-glucoside |
| − | CAS_RN 98568-80-0 | + | CAS_RN 98568-80-0 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES O(C=1c(c5)ccc(c5O)O)c(c2)c(c(c(C(O3)C(C(O)C(O)C(COC(C4O)OC(C(C(O)4)O)CO)3)O)c2O)O)C(C1)=O | + | SMILES O(C=1c(c5)ccc(c5O)O)c(c2)c(c(c(C(O3)C(C(O)C(O)C(COC(C4O)OC(C(C(O)4)O)CO)3)O)c2O)O)C(C1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
0.6849 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6849 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2412 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7975 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7975 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2412 0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3538 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9101 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9101 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3538 0.3725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3538 -1.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2412 -1.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5900 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1762 0.1371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7624 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7624 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1762 1.4909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5900 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4740 1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8685 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3529 -1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8372 -1.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1360 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6310 -0.7324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1879 -1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3803 -1.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9952 -1.4955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1126 0.3818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5279 -1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7779 -0.4104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9622 0.1132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4466 -0.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9310 -0.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2297 -0.2374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7247 0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2816 -0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4740 -0.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0890 -0.5468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6216 -0.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6636 0.3287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3482 0.1373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6346 0.3345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2180 0.9179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
25 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
40 30 1 0 0 0 0
15 41 1 0 0 0 0
36 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 ^CH2OH
M SBV 1 46 -7.2746 5.6558
S SKP 8
ID FL3FACCS0030
KNApSAcK_ID C00006211
NAME Isoorientin 6''-O-glucoside
CAS_RN 98568-80-0
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES O(C=1c(c5)ccc(c5O)O)c(c2)c(c(c(C(O3)C(C(O)C(O)C(COC(C4O)OC(C(C(O)4)O)CO)3)O)c2O)O)C(C1)=O
M END