Mol:FL3FACCS0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3049 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3049 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3049 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3049 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5901 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5901 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1246 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1246 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1246 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1246 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5901 1.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5901 1.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8394 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8394 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5541 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5541 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5541 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5541 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8394 1.2416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8394 1.2416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8394 -1.0525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8394 -1.0525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9219 1.4460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9219 1.4460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5901 -1.2341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5901 -1.2341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3482 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3482 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1014 0.8331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1014 0.8331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8545 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8545 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8545 2.1378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8545 2.1378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1014 2.5726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1014 2.5726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3482 2.1378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3482 2.1378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6073 2.5723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6073 2.5723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6943 0.0101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6943 0.0101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2495 -0.5772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2495 -0.5772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6088 -0.3280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6088 -0.3280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9411 -0.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9411 -0.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4399 0.1279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4399 0.1279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0943 -0.1071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0943 -0.1071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1903 -0.2373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1903 -0.2373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9429 -0.6110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9429 -0.6110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9929 -0.7394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9929 -0.7394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0874 -1.4275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0874 -1.4275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4218 -2.0067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4218 -2.0067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7787 -1.8230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7787 -1.8230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1316 -2.0067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1316 -2.0067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7970 -1.4275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7970 -1.4275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4402 -1.6113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4402 -1.6113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7085 -1.5939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7085 -1.5939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8627 -1.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8627 -1.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1465 -2.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1465 -2.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1302 -2.5726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1302 -2.5726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6073 0.8334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6073 0.8334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5834 0.4035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5834 0.4035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6073 0.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6073 0.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 14 9 1 0 0 0 0 | + | 14 9 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 46 0.4892 -0.5105 | + | M SBV 1 46 0.4892 -0.5105 |
| − | S SKP 5 | + | S SKP 5 |
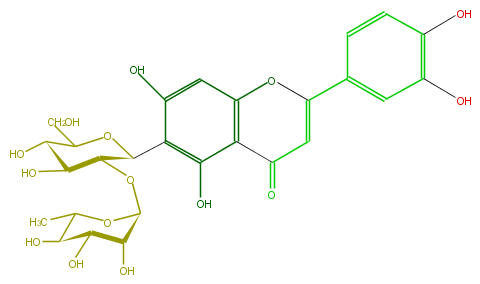
| − | ID FL3FACCS0027 | + | ID FL3FACCS0027 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(c1)c(c(cc1C(=C5)Oc(c4)c(C5=O)c(c(c4O)C(C(OC(C(O)3)OC(C)C(C(O)3)O)2)OC(C(O)C2O)CO)O)O)O | + | SMILES c(c1)c(c(cc1C(=C5)Oc(c4)c(C5=O)c(c(c4O)C(C(OC(C(O)3)OC(C)C(C(O)3)O)2)OC(C(O)C2O)CO)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.3049 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3049 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5901 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1246 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1246 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5901 1.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8394 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5541 0.0036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5541 0.8290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8394 1.2416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8394 -1.0525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9219 1.4460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5901 -1.2341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3482 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1014 0.8331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8545 1.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8545 2.1378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1014 2.5726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3482 2.1378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6073 2.5723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6943 0.0101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2495 -0.5772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6088 -0.3280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9411 -0.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4399 0.1279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0943 -0.1071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1903 -0.2373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9429 -0.6110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9929 -0.7394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0874 -1.4275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4218 -2.0067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7787 -1.8230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1316 -2.0067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7970 -1.4275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4402 -1.6113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7085 -1.5939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8627 -1.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1465 -2.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1302 -2.5726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6073 0.8334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5834 0.4035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6073 0.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
14 9 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 29 1 0 0 0 0
24 2 1 0 0 0 0
16 40 1 0 0 0 0
41 42 1 0 0 0 0
26 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^CH2OH
M SBV 1 46 0.4892 -0.5105
S SKP 5
ID FL3FACCS0027
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(c1)c(c(cc1C(=C5)Oc(c4)c(C5=O)c(c(c4O)C(C(OC(C(O)3)OC(C)C(C(O)3)O)2)OC(C(O)C2O)CO)O)O)O
M END