Mol:FL3FACCS0019
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1018 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1018 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1018 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1018 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3874 -2.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3874 -2.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3271 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3271 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3271 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3271 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3874 -0.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3874 -0.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0415 -2.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0415 -2.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7559 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7559 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7559 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7559 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0415 -0.5937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0415 -0.5937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0415 -2.8869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0415 -2.8869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8160 -0.5938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8160 -0.5938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2372 2.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2372 2.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0150 1.7903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0150 1.7903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6850 0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6850 0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6762 0.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6762 0.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0812 0.7180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0812 0.7180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4729 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4729 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6349 3.0683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6349 3.0683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0150 2.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0150 2.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0779 0.2613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0779 0.2613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3874 -3.0683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3874 -3.0683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5496 -0.5674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5496 -0.5674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3024 -1.0021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3024 -1.0021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0552 -0.5674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0552 -0.5674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0552 0.3019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0552 0.3019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3024 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3024 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5496 0.3019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5496 0.3019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8077 0.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8077 0.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7689 -2.0431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7689 -2.0431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2923 -2.6723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2923 -2.6723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6059 -2.4054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6059 -2.4054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9435 -2.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9435 -2.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4248 -1.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4248 -1.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0253 -2.2337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0253 -2.2337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3693 -2.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3693 -2.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9064 -2.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9064 -2.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0407 -2.7317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0407 -2.7317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8077 -1.0018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8077 -1.0018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5815 -1.6776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5815 -1.6776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8077 -1.2960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8077 -1.2960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.5561 -0.5561 | + | M SBV 1 45 0.5561 -0.5561 |
| − | S SKP 5 | + | S SKP 5 |
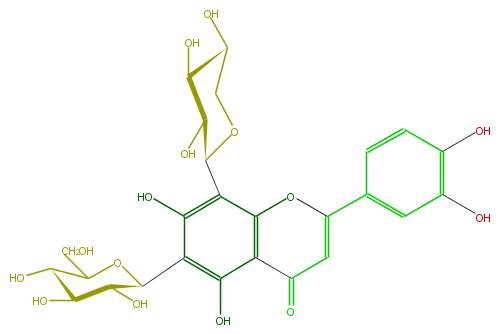
| − | ID FL3FACCS0019 | + | ID FL3FACCS0019 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O | + | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.1018 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1018 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3874 -2.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3271 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3271 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3874 -0.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0415 -2.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7559 -1.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7559 -1.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0415 -0.5937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0415 -2.8869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8160 -0.5938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2372 2.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0150 1.7903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6850 0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6762 0.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0812 0.7180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4729 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6349 3.0683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0150 2.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0779 0.2613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3874 -3.0683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5496 -0.5674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3024 -1.0021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0552 -0.5674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0552 0.3019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3024 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5496 0.3019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8077 0.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7689 -2.0431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2923 -2.6723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6059 -2.4054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9435 -2.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4248 -1.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0253 -2.2337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3693 -2.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9064 -2.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0407 -2.7317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8077 -1.0018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5815 -1.6776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8077 -1.2960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
25 39 1 0 0 0 0
40 41 1 0 0 0 0
35 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.5561 -0.5561
S SKP 5
ID FL3FACCS0019
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O
M END