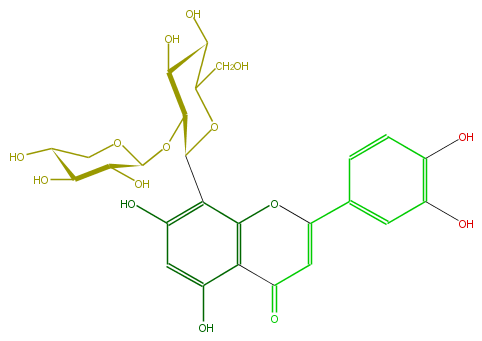
Mol:FL3FACCS0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4905 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4905 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4905 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4905 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7761 -2.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7761 -2.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0616 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0616 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0616 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0616 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7761 -0.6621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7761 -0.6621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6528 -2.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6528 -2.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6528 -0.6621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6528 -0.6621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6528 -2.9553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6528 -2.9553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2047 -0.6622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2047 -0.6622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6878 2.5378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6878 2.5378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4655 1.9487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4655 1.9487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1356 1.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1356 1.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1267 0.2817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1267 0.2817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5317 0.8765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5317 0.8765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9234 1.6187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9234 1.6187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0336 3.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0336 3.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4655 2.6273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4655 2.6273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5043 0.4797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5043 0.4797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7761 -3.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7761 -3.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1609 -0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1609 -0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9137 -1.0705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9137 -1.0705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6665 -0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6665 -0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6665 0.2335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6665 0.2335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9137 0.6681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9137 0.6681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1609 0.2335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1609 0.2335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4190 0.6679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4190 0.6679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8183 0.4364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8183 0.4364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3417 -0.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3417 -0.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6553 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6553 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9929 0.0812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9929 0.0812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4742 0.5626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4742 0.5626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0747 0.2457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0747 0.2457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4190 0.2755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4190 0.2755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9388 -0.1929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9388 -0.1929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0604 -0.2693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0604 -0.2693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4190 -1.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4190 -1.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4418 2.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4418 2.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4757 1.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4757 1.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 21 1 0 0 0 0 | + | 33 21 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 18 40 1 0 0 0 0 | + | 18 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.4816 -0.4816 | + | M SBV 1 45 -0.4816 -0.4816 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACCS0014 | + | ID FL3FACCS0014 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES OC(C(O)1)C(O)COC(OC(C5O)C(OC(C(O)5)CO)c(c4O)c(c(c(c4)O)2)OC(c(c3)ccc(O)c3O)=CC2=O)1 | + | SMILES OC(C(O)1)C(O)COC(OC(C5O)C(OC(C(O)5)CO)c(c4O)c(c(c(c4)O)2)OC(c(c3)ccc(O)c3O)=CC2=O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.4905 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4905 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7761 -2.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0616 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0616 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7761 -0.6621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6528 -2.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 -1.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 -1.0746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6528 -0.6621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6528 -2.9553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2047 -0.6622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6878 2.5378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4655 1.9487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1356 1.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1267 0.2817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5317 0.8765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9234 1.6187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0336 3.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4655 2.6273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5043 0.4797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7761 -3.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1609 -0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9137 -1.0705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6665 -0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6665 0.2335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9137 0.6681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1609 0.2335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4190 0.6679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8183 0.4364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3417 -0.1929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6553 0.0741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9929 0.0812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4742 0.5626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0747 0.2457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4190 0.2755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9388 -0.1929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0604 -0.2693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4190 -1.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4418 2.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4757 1.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 21 1 0 0 0 0
25 39 1 0 0 0 0
40 41 1 0 0 0 0
18 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.4816 -0.4816
S SKP 5
ID FL3FACCS0014
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES OC(C(O)1)C(O)COC(OC(C5O)C(OC(C(O)5)CO)c(c4O)c(c(c(c4)O)2)OC(c(c3)ccc(O)c3O)=CC2=O)1
M END