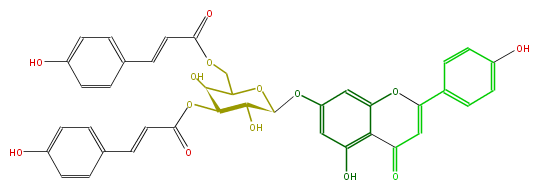
Mol:FL3FAAGS0068
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.5403 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5403 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5403 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5403 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2548 -1.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2548 -1.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9693 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9693 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9693 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9693 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2548 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2548 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6837 -1.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6837 -1.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3982 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3982 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3982 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3982 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6837 0.1112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6837 0.1112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1127 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1127 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8271 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8271 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5416 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5416 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5416 0.9362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5416 0.9362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8271 1.3487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8271 1.3487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1127 0.9362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1127 0.9362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6837 -2.3638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6837 -2.3638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2548 -2.3638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2548 -2.3638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2879 1.3671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.2879 1.3671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8207 0.0457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8207 0.0457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8573 0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8573 0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4446 -0.4930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4446 -0.4930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6463 -0.2838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6463 -0.2838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1472 -0.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1472 -0.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2654 0.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2654 0.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0638 -0.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0638 -0.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2321 0.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2321 0.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2049 0.5694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2049 0.5694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3100 -0.2611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3100 -0.2611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4766 -0.9171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4766 -0.9171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7484 0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7484 0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1603 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1603 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7484 2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7484 2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9842 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9842 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3961 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3961 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2200 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2200 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6325 1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6325 1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4575 1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4575 1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8700 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8700 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4575 0.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4575 0.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6325 0.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6325 0.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6939 0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6939 0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7543 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7543 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3498 -1.6552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3498 -1.6552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5782 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5782 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9901 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9901 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8140 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8140 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2265 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2265 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0515 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0515 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4640 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4640 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0515 -2.3825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0515 -2.3825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2265 -2.3825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2265 -2.3825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.2879 -1.6680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.2879 -1.6680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 31 27 1 0 0 0 0 | + | 31 27 1 0 0 0 0 |
| − | 29 43 1 0 0 0 0 | + | 29 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0068 | + | ID FL3FAAGS0068 |
| − | KNApSAcK_ID C00013618 | + | KNApSAcK_ID C00013618 |
| − | NAME Apigenin 7-(3'',6''-Di-E-p-coumaroylgalactoside);7-[[3,6-Bis-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-b-D-galactopyranosyl]oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one | + | NAME Apigenin 7-(3'',6''-Di-E-p-coumaroylgalactoside);7-[[3,6-Bis-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-b-D-galactopyranosyl]oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 480990-58-7 | + | CAS_RN 480990-58-7 |
| − | FORMULA C39H32O14 | + | FORMULA C39H32O14 |
| − | EXACTMASS 724.179205732 | + | EXACTMASS 724.179205732 |
| − | AVERAGEMASS 724.6629800000001 | + | AVERAGEMASS 724.6629800000001 |
| − | SMILES Oc(c6)ccc(c6)C(=C5)Oc(c1)c(C5=O)c(cc(OC(C2O)OC(COC(C=Cc(c4)ccc(c4)O)=O)C(C2OC(=O)C=Cc(c3)ccc(O)c3)O)1)O | + | SMILES Oc(c6)ccc(c6)C(=C5)Oc(c1)c(C5=O)c(cc(OC(C2O)OC(COC(C=Cc(c4)ccc(c4)O)=O)C(C2OC(=O)C=Cc(c3)ccc(O)c3)O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
1.5403 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5403 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2548 -1.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9693 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9693 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2548 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6837 -1.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3982 -1.1263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3982 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6837 0.1112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1127 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8271 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5416 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5416 0.9362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8271 1.3487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1127 0.9362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6837 -2.3638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2548 -2.3638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.2879 1.3671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8207 0.0457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8573 0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4446 -0.4930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6463 -0.2838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1472 -0.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2654 0.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0638 -0.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2321 0.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2049 0.5694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3100 -0.2611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4766 -0.9171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7484 0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1603 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7484 2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9842 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3961 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2200 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6325 1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4575 1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8700 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4575 0.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6325 0.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6939 0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7543 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3498 -1.6552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5782 -0.9545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9901 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8140 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2265 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0515 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.4640 -1.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0515 -2.3825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2265 -2.3825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.2879 -1.6680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
3 18 1 0 0 0 0
14 19 1 0 0 0 0
20 1 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 20 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
31 27 1 0 0 0 0
29 43 1 0 0 0 0
S SKP 8
ID FL3FAAGS0068
KNApSAcK_ID C00013618
NAME Apigenin 7-(3'',6''-Di-E-p-coumaroylgalactoside);7-[[3,6-Bis-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-b-D-galactopyranosyl]oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 480990-58-7
FORMULA C39H32O14
EXACTMASS 724.179205732
AVERAGEMASS 724.6629800000001
SMILES Oc(c6)ccc(c6)C(=C5)Oc(c1)c(C5=O)c(cc(OC(C2O)OC(COC(C=Cc(c4)ccc(c4)O)=O)C(C2OC(=O)C=Cc(c3)ccc(O)c3)O)1)O
M END