Mol:FL3FAADS0045
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.0997 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0997 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3852 0.1690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3852 0.1690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6707 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6707 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6707 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6707 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3852 -1.4810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3852 -1.4810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0997 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0997 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9562 0.1690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9562 0.1690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2418 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2418 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2418 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2418 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9562 -1.4810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9562 -1.4810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3852 -2.0531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3852 -2.0531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9010 0.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9010 0.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6155 -0.1933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6155 -0.1933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3300 0.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3300 0.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3300 1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3300 1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6155 1.4567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6155 1.4567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9010 1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9010 1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.0134 1.4388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.0134 1.4388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9562 -2.2510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9562 -2.2510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5513 0.1551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5513 0.1551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1247 -1.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1247 -1.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7121 -2.0302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7121 -2.0302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9137 -1.8211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9137 -1.8211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1202 -2.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1202 -2.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5328 -1.3332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5328 -1.3332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3312 -1.5423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3312 -1.5423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4882 -0.9564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4882 -0.9564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9680 -0.6794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9680 -0.6794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6079 -1.7988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6079 -1.7988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1607 -1.7712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1607 -1.7712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0852 -2.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0852 -2.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1247 0.3180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1247 0.3180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7121 -0.3967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7121 -0.3967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9137 -0.1876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9137 -0.1876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1202 -0.4145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1202 -0.4145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5328 0.3003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5328 0.3003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3312 0.0912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3312 0.0912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4882 0.6771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4882 0.6771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9680 0.9541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9680 0.9541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6079 -0.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6079 -0.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1607 -0.1377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1607 -0.1377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0852 -0.8275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0852 -0.8275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4119 1.7475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4119 1.7475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9999 2.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9999 2.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2358 1.7475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2358 1.7475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6477 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6477 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4716 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4716 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8841 1.7485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8841 1.7485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7091 1.7485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7091 1.7485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1216 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1216 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7091 0.3196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7091 0.3196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8841 0.3196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8841 0.3196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.9455 1.0340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.9455 1.0340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2873 2.3267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2873 2.3267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.0134 2.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.0134 2.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 9 1 0 0 0 0 | + | 24 9 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 49 54 1 0 0 0 0 | + | 49 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 20 35 1 0 0 0 0 | + | 20 35 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAADS0045 | + | ID FL3FAADS0045 |
| − | KNApSAcK_ID C00014078 | + | KNApSAcK_ID C00014078 |
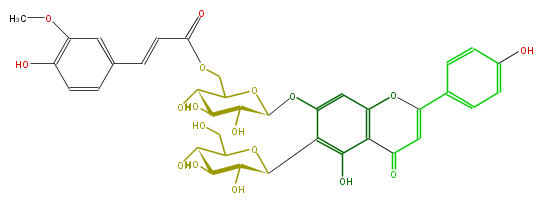
| − | NAME Isovitexin 7-O-beta-[6'''-O-(E)-p-feruloyl]glucoside;6'''-Feruloylsaponarin | + | NAME Isovitexin 7-O-beta-[6'''-O-(E)-p-feruloyl]glucoside;6'''-Feruloylsaponarin |
| − | CAS_RN 212271-12-0 | + | CAS_RN 212271-12-0 |
| − | FORMULA C37H38O18 | + | FORMULA C37H38O18 |
| − | EXACTMASS 770.205814412 | + | EXACTMASS 770.205814412 |
| − | AVERAGEMASS 770.6868199999999 | + | AVERAGEMASS 770.6868199999999 |
| − | SMILES COc(c(O)6)cc(cc6)C=CC(OCC(C(O)1)OC(Oc(c(C(C(O)5)OC(C(C(O)5)O)CO)2)cc(O3)c(C(C=C3c(c4)ccc(O)c4)=O)c(O)2)C(O)C(O)1)=O | + | SMILES COc(c(O)6)cc(cc6)C=CC(OCC(C(O)1)OC(Oc(c(C(C(O)5)OC(C(C(O)5)O)CO)2)cc(O3)c(C(C=C3c(c4)ccc(O)c4)=O)c(O)2)C(O)C(O)1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
4.0997 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3852 0.1690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6707 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6707 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3852 -1.4810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0997 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9562 0.1690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2418 -0.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2418 -1.0685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9562 -1.4810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3852 -2.0531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9010 0.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6155 -0.1933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3300 0.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3300 1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6155 1.4567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9010 1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.0134 1.4388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9562 -2.2510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5513 0.1551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1247 -1.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7121 -2.0302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9137 -1.8211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1202 -2.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5328 -1.3332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3312 -1.5423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4882 -0.9564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9680 -0.6794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6079 -1.7988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1607 -1.7712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0852 -2.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1247 0.3180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7121 -0.3967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9137 -0.1876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1202 -0.4145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5328 0.3003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3312 0.0912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4882 0.6771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9680 0.9541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6079 -0.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1607 -0.1377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0852 -0.8275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4119 1.7475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9999 2.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2358 1.7475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6477 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4716 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8841 1.7485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7091 1.7485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1216 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7091 0.3196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8841 0.3196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.9455 1.0340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.2873 2.3267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.0134 2.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
1 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
10 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
21 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 9 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
49 54 1 0 0 0 0
54 55 1 0 0 0 0
20 35 1 0 0 0 0
39 43 1 0 0 0 0
S SKP 8
ID FL3FAADS0045
KNApSAcK_ID C00014078
NAME Isovitexin 7-O-beta-[6'''-O-(E)-p-feruloyl]glucoside;6'''-Feruloylsaponarin
CAS_RN 212271-12-0
FORMULA C37H38O18
EXACTMASS 770.205814412
AVERAGEMASS 770.6868199999999
SMILES COc(c(O)6)cc(cc6)C=CC(OCC(C(O)1)OC(Oc(c(C(C(O)5)OC(C(C(O)5)O)CO)2)cc(O3)c(C(C=C3c(c4)ccc(O)c4)=O)c(O)2)C(O)C(O)1)=O
M END