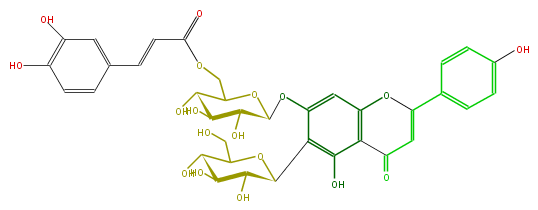
Mol:FL3FAADS0043
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.9265 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9265 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2121 0.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2121 0.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4976 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4976 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4976 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4976 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2121 -1.3820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2121 -1.3820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9265 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9265 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7831 0.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7831 0.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0687 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0687 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0687 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0687 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7831 -1.3820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7831 -1.3820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2121 -1.9541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2121 -1.9541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8358 -1.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8358 -1.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4232 -2.0797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4232 -2.0797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6248 -1.8706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6248 -1.8706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1686 -2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1686 -2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2439 -1.3827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2439 -1.3827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0423 -1.5917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0423 -1.5917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1993 -1.0059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1993 -1.0059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6791 -0.7289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6791 -0.7289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3190 -1.8482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3190 -1.8482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8719 -1.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8719 -1.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7963 -2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7963 -2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7279 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7279 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4424 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4424 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1569 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1569 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1569 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1569 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4424 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4424 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7279 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7279 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.8403 1.5378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.8403 1.5378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7831 -2.1520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7831 -2.1520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3782 0.2542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3782 0.2542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9843 0.3345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9843 0.3345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5717 -0.3802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5717 -0.3802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7733 -0.1711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7733 -0.1711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0201 -0.3979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0201 -0.3979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3924 0.3168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3924 0.3168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1908 0.1078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1908 0.1078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3478 0.6936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3478 0.6936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4675 -0.1487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4675 -0.1487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0204 -0.1212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0204 -0.1212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9448 -0.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9448 -0.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8948 1.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8948 1.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3067 1.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3067 1.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8948 2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8948 2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1306 1.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1306 1.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5425 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5425 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3664 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3664 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7789 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7789 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6039 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6039 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0164 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0164 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6039 0.3690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6039 0.3690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7789 0.3690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7789 0.3690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.8403 1.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.8403 1.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1592 2.3533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1592 2.3533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 12 13 1 1 0 0 0 | + | 12 13 1 1 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 15 14 1 1 0 0 0 | + | 15 14 1 1 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 12 20 1 0 0 0 0 | + | 12 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 9 15 1 0 0 0 0 | + | 9 15 1 0 0 0 0 |
| − | 1 23 1 0 0 0 0 | + | 1 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 10 30 1 0 0 0 0 | + | 10 30 1 0 0 0 0 |
| − | 8 31 1 0 0 0 0 | + | 8 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 42 38 1 0 0 0 0 | + | 42 38 1 0 0 0 0 |
| − | 49 54 1 0 0 0 0 | + | 49 54 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAADS0043 | + | ID FL3FAADS0043 |
| − | KNApSAcK_ID C00014076 | + | KNApSAcK_ID C00014076 |
| − | NAME Isovitexin 7-O-(6'''-caffeoyl)-beta-D-glucopyranoside | + | NAME Isovitexin 7-O-(6'''-caffeoyl)-beta-D-glucopyranoside |
| − | CAS_RN 165746-63-4 | + | CAS_RN 165746-63-4 |
| − | FORMULA C36H36O18 | + | FORMULA C36H36O18 |
| − | EXACTMASS 756.190164348 | + | EXACTMASS 756.190164348 |
| − | AVERAGEMASS 756.6602399999999 | + | AVERAGEMASS 756.6602399999999 |
| − | SMILES Oc(c3C(C(O)6)OC(C(C(O)6)O)CO)c(c2cc(OC(O4)C(O)C(O)C(O)C4COC(C=Cc(c5)ccc(c(O)5)O)=O)3)C(C=C(O2)c(c1)ccc(O)c1)=O | + | SMILES Oc(c3C(C(O)6)OC(C(C(O)6)O)CO)c(c2cc(OC(O4)C(O)C(O)C(O)C4COC(C=Cc(c5)ccc(c(O)5)O)=O)3)C(C=C(O2)c(c1)ccc(O)c1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
3.9265 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2121 0.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4976 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4976 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2121 -1.3820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9265 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7831 0.2680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0687 -0.1445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0687 -0.9695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7831 -1.3820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2121 -1.9541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8358 -1.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4232 -2.0797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6248 -1.8706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1686 -2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2439 -1.3827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0423 -1.5917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1993 -1.0059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6791 -0.7289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3190 -1.8482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8719 -1.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7963 -2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7279 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4424 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1569 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1569 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4424 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7279 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.8403 1.5378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7831 -2.1520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3782 0.2542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9843 0.3345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5717 -0.3802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7733 -0.1711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0201 -0.3979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3924 0.3168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1908 0.1078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3478 0.6936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4675 -0.1487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0204 -0.1212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9448 -0.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8948 1.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3067 1.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8948 2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1306 1.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5425 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3664 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7789 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6039 1.7980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0164 1.0835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6039 0.3690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7789 0.3690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.8403 1.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.1592 2.3533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
12 13 1 1 0 0 0
13 14 1 1 0 0 0
15 14 1 1 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 12 1 0 0 0 0
17 18 1 0 0 0 0
18 19 1 0 0 0 0
12 20 1 0 0 0 0
13 21 1 0 0 0 0
14 22 1 0 0 0 0
9 15 1 0 0 0 0
1 23 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
26 29 1 0 0 0 0
10 30 1 0 0 0 0
8 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
34 41 1 0 0 0 0
35 31 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
42 38 1 0 0 0 0
49 54 1 0 0 0 0
S SKP 8
ID FL3FAADS0043
KNApSAcK_ID C00014076
NAME Isovitexin 7-O-(6'''-caffeoyl)-beta-D-glucopyranoside
CAS_RN 165746-63-4
FORMULA C36H36O18
EXACTMASS 756.190164348
AVERAGEMASS 756.6602399999999
SMILES Oc(c3C(C(O)6)OC(C(C(O)6)O)CO)c(c2cc(OC(O4)C(O)C(O)C(O)C4COC(C=Cc(c5)ccc(c(O)5)O)=O)3)C(C=C(O2)c(c1)ccc(O)c1)=O
M END