Mol:FL3FAACS0081
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.5611 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5611 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8466 0.9517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8466 0.9517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1321 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1321 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1321 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1321 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8466 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8466 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5611 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5611 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4177 0.9517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4177 0.9517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7032 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7032 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7032 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7032 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4177 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4177 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8466 -1.2704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8466 -1.2704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3625 1.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3625 1.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0769 0.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0769 0.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.7914 1.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.7914 1.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.7914 1.8269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.7914 1.8269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0769 2.2394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0769 2.2394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3625 1.8269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3625 1.8269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.4749 2.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.4749 2.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4177 -1.4683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4177 -1.4683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0127 0.9378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0127 0.9378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0363 0.0281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0363 0.0281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6236 -0.6866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6236 -0.6866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1747 -0.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1747 -0.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9682 -0.7043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9682 -0.7043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5556 0.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5556 0.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2428 -0.1986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2428 -0.1986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3997 0.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3997 0.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5195 -0.4551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5195 -0.4551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0723 -0.4275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0723 -0.4275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0032 -1.1174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0032 -1.1174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8606 0.6533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8606 0.6533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6368 -1.0939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6368 -1.0939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2241 -1.8086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2241 -1.8086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4258 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4258 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6323 -1.8263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6323 -1.8263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0449 -1.1115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0449 -1.1115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8433 -1.3206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8433 -1.3206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0002 -0.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0002 -0.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4801 -0.4577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4801 -0.4577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1200 -1.5771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1200 -1.5771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6728 -1.5495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6728 -1.5495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5973 -2.2394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5973 -2.2394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9413 0.2711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9413 0.2711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5293 0.9846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5293 0.9846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7651 0.2711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7651 0.2711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1771 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1771 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0010 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0010 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4135 0.2720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4135 0.2720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2385 0.2720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2385 0.2720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6510 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6510 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2385 -1.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2385 -1.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4135 -1.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4135 -1.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.4749 -0.4424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.4749 -0.4424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 9 1 0 0 0 0 | + | 24 9 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAACS0081 | + | ID FL3FAACS0081 |
| − | KNApSAcK_ID C00014080 | + | KNApSAcK_ID C00014080 |
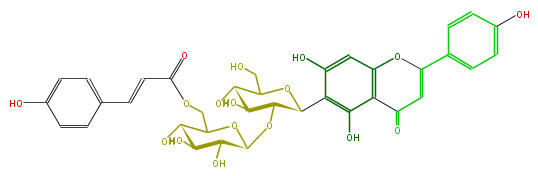
| − | NAME Isovitexin 2''-O-(6'''-(E)-p-coumaroyl)glucoside | + | NAME Isovitexin 2''-O-(6'''-(E)-p-coumaroyl)glucoside |
| − | CAS_RN 220948-76-5 | + | CAS_RN 220948-76-5 |
| − | FORMULA C36H36O17 | + | FORMULA C36H36O17 |
| − | EXACTMASS 740.1952497259999 | + | EXACTMASS 740.1952497259999 |
| − | AVERAGEMASS 740.66084 | + | AVERAGEMASS 740.66084 |
| − | SMILES OCC(C6O)OC(C(C6O)OC(O4)C(O)C(C(C4COC(=O)C=Cc(c5)ccc(c5)O)O)O)c(c1O)c(O)cc(O2)c(C(=O)C=C(c(c3)ccc(O)c3)2)1 | + | SMILES OCC(C6O)OC(C(C6O)OC(O4)C(O)C(C(C4COC(=O)C=Cc(c5)ccc(c5)O)O)O)c(c1O)c(O)cc(O2)c(C(=O)C=C(c(c3)ccc(O)c3)2)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
4.5611 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8466 0.9517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1321 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1321 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8466 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5611 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4177 0.9517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7032 0.5392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7032 -0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4177 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8466 -1.2704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3625 1.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0769 0.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.7914 1.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.7914 1.8269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0769 2.2394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3625 1.8269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.4749 2.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4177 -1.4683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0127 0.9378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0363 0.0281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6236 -0.6866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1747 -0.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9682 -0.7043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5556 0.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2428 -0.1986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3997 0.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5195 -0.4551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0723 -0.4275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0032 -1.1174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8606 0.6533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6368 -1.0939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2241 -1.8086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4258 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6323 -1.8263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0449 -1.1115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8433 -1.3206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0002 -0.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4801 -0.4577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1200 -1.5771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6728 -1.5495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5973 -2.2394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9413 0.2711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5293 0.9846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7651 0.2711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1771 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0010 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4135 0.2720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2385 0.2720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6510 -0.4424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2385 -1.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4135 -1.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.4749 -0.4424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
1 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
10 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
27 31 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 9 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
35 30 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
39 43 1 0 0 0 0
S SKP 8
ID FL3FAACS0081
KNApSAcK_ID C00014080
NAME Isovitexin 2''-O-(6'''-(E)-p-coumaroyl)glucoside
CAS_RN 220948-76-5
FORMULA C36H36O17
EXACTMASS 740.1952497259999
AVERAGEMASS 740.66084
SMILES OCC(C6O)OC(C(C6O)OC(O4)C(O)C(C(C4COC(=O)C=Cc(c5)ccc(c5)O)O)O)c(c1O)c(O)cc(O2)c(C(=O)C=C(c(c3)ccc(O)c3)2)1
M END