Mol:FL3FAACS0028
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4199 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4199 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4199 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4199 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8636 -1.8623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8636 -1.8623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3073 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3073 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3073 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3073 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8636 -0.5776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8636 -0.5776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7509 -1.8623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7509 -1.8623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1946 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1946 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1946 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1946 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7509 -0.5776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7509 -0.5776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7509 -2.3632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7509 -2.3632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9760 -0.5777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9760 -0.5777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4233 -0.5571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4233 -0.5571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0095 -0.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0095 -0.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5958 -0.5571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5958 -0.5571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5958 0.1198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5958 0.1198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0095 0.4582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0095 0.4582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4233 0.1198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4233 0.1198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1816 0.4580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1816 0.4580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6388 1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6388 1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2444 1.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2444 1.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9874 0.7812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9874 0.7812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9806 0.1437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9806 0.1437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5172 0.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5172 0.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8223 1.1849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8223 1.1849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8006 2.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8006 2.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2792 2.1570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2792 2.1570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6229 0.4026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6229 0.4026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8636 -2.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8636 -2.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0489 -1.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0489 -1.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7481 -1.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7481 -1.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3265 -1.7400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3265 -1.7400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9085 -1.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9085 -1.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2094 -1.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2094 -1.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6309 -1.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6309 -1.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4902 -1.5340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4902 -1.5340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8691 -1.1370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8691 -1.1370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6435 -1.6349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6435 -1.6349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2615 -1.8786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2615 -1.8786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9760 -2.2911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9760 -2.2911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3069 1.4824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3069 1.4824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5925 1.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5925 1.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 9 1 0 0 0 0 | + | 13 9 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 31 8 1 0 0 0 0 | + | 31 8 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 43 -7.1159 5.7279 | + | M SBV 1 43 -7.1159 5.7279 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 45 -6.9535 5.9987 | + | M SBV 2 45 -6.9535 5.9987 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAACS0028 | + | ID FL3FAACS0028 |
| − | KNApSAcK_ID C00006233 | + | KNApSAcK_ID C00006233 |
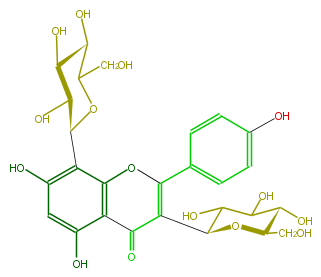
| − | NAME 3,8-Di-C-glucopyranosylapigenin | + | NAME 3,8-Di-C-glucopyranosylapigenin |
| − | CAS_RN 97218-31-0 | + | CAS_RN 97218-31-0 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(c1O)cc(C(=C(C(O5)C(C(O)C(O)C5CO)O)2)Oc(c(C(C(O)4)OC(C(O)C(O)4)CO)3)c(c(O)cc3O)C(=O)2)cc1 | + | SMILES c(c1O)cc(C(=C(C(O5)C(C(O)C(O)C5CO)O)2)Oc(c(C(C(O)4)OC(C(O)C(O)4)CO)3)c(c(O)cc3O)C(=O)2)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.4199 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4199 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8636 -1.8623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3073 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3073 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8636 -0.5776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7509 -1.8623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1946 -1.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1946 -0.8988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7509 -0.5776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7509 -2.3632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9760 -0.5777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4233 -0.5571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0095 -0.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5958 -0.5571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5958 0.1198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0095 0.4582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4233 0.1198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1816 0.4580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6388 1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2444 1.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9874 0.7812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9806 0.1437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5172 0.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8223 1.1849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8006 2.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2792 2.1570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6229 0.4026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8636 -2.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0489 -1.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7481 -1.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3265 -1.7400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9085 -1.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2094 -1.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6309 -1.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4902 -1.5340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8691 -1.1370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6435 -1.6349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2615 -1.8786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9760 -2.2911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3069 1.4824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5925 1.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
13 9 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
6 23 1 0 0 0 0
3 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
31 8 1 0 0 0 0
33 39 1 0 0 0 0
39 40 1 0 0 0 0
25 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SBV 1 43 -7.1159 5.7279
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SBV 2 45 -6.9535 5.9987
S SKP 8
ID FL3FAACS0028
KNApSAcK_ID C00006233
NAME 3,8-Di-C-glucopyranosylapigenin
CAS_RN 97218-31-0
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(c1O)cc(C(=C(C(O5)C(C(O)C(O)C5CO)O)2)Oc(c(C(C(O)4)OC(C(O)C(O)4)CO)3)c(c(O)cc3O)C(=O)2)cc1
M END