Mol:FL3FAACS0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1653 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1653 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1653 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1653 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6090 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6090 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0527 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0527 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0527 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0527 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6090 0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6090 0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5036 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5036 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0599 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0599 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0599 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0599 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5036 0.3725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5036 0.3725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5036 -1.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5036 -1.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6090 -1.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6090 -1.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7397 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7397 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3260 0.1371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3260 0.1371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9122 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9122 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9122 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9122 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3260 1.4909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3260 1.4909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7397 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7397 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6238 1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6238 1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7187 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7187 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2031 -1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2031 -1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6874 -1.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6874 -1.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9862 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9862 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4812 -0.7324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4812 -0.7324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0381 -1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0381 -1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2305 -1.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2305 -1.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8454 -1.4955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8454 -1.4955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7376 0.3818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7376 0.3818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3781 -1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3781 -1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3034 -0.4960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3034 -0.4960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0026 -1.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0026 -1.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5811 -0.8517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5811 -0.8517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1630 -1.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1630 -1.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4639 -0.4960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4639 -0.4960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8854 -0.6613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8854 -0.6613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7447 -0.6457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7447 -0.6457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1236 -0.2487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1236 -0.2487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8980 -0.7466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8980 -0.7466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5160 -0.9903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5160 -0.9903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2305 -1.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2305 -1.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3911 -0.6142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3911 -0.6142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9744 -0.0309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9744 -0.0309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 31 8 1 0 0 0 0 | + | 31 8 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 43 -4.5207 9.0293 | + | M SBV 1 43 -4.5207 9.0293 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 45 -5.2267 9.3889 | + | M SBV 2 45 -5.2267 9.3889 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAACS0027 | + | ID FL3FAACS0027 |
| − | KNApSAcK_ID C00006232 | + | KNApSAcK_ID C00006232 |
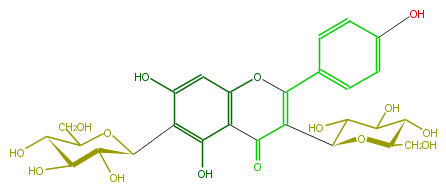
| − | NAME 3,6-Di-C-glucosylapigenin | + | NAME 3,6-Di-C-glucosylapigenin |
| − | CAS_RN 90456-55-6 | + | CAS_RN 90456-55-6 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(C(O)1)(O)C(O)C(C(C(=O)3)=C(Oc(c4)c3c(c(C(O5)C(C(O)C(O)C5CO)O)c(O)4)O)c(c2)ccc(O)c2)OC(CO)1 | + | SMILES C(C(O)1)(O)C(O)C(C(C(=O)3)=C(Oc(c4)c3c(c(C(O5)C(C(O)C(O)C5CO)O)c(O)4)O)c(c2)ccc(O)c2)OC(CO)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.1653 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1653 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6090 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0527 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0527 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6090 0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5036 -0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0599 -0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0599 0.0514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5036 0.3725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5036 -1.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6090 -1.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7397 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3260 0.1371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9122 0.4756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9122 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3260 1.4909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7397 1.1525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6238 1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7187 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2031 -1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6874 -1.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9862 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4812 -0.7324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0381 -1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2305 -1.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8454 -1.4955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7376 0.3818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3781 -1.6395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3034 -0.4960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0026 -1.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5811 -0.8517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1630 -1.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4639 -0.4960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8854 -0.6613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7447 -0.6457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1236 -0.2487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8980 -0.7466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5160 -0.9903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2305 -1.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3911 -0.6142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9744 -0.0309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
31 8 1 0 0 0 0
33 39 1 0 0 0 0
39 40 1 0 0 0 0
25 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SBV 1 43 -4.5207 9.0293
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 ^CH2OH
M SBV 2 45 -5.2267 9.3889
S SKP 8
ID FL3FAACS0027
KNApSAcK_ID C00006232
NAME 3,6-Di-C-glucosylapigenin
CAS_RN 90456-55-6
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(C(O)1)(O)C(O)C(C(C(=O)3)=C(Oc(c4)c3c(c(C(O5)C(C(O)C(O)C5CO)O)c(O)4)O)c(c2)ccc(O)c2)OC(CO)1
M END