Mol:FL1C3AGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 47 0 0 0 0 0 0 0 0999 V2000 | + | 44 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.8692 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8692 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8692 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8692 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5837 -1.4819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5837 -1.4819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2981 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2981 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2981 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2981 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5837 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5837 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0126 -1.4819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0126 -1.4819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7271 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7271 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7271 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7271 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4416 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4416 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1560 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1560 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8705 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8705 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8705 0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8705 0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1560 1.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1560 1.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4416 0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4416 0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0126 -2.3069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0126 -2.3069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5590 1.3906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5590 1.3906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4542 0.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4542 0.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9763 -0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9763 -0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2011 -0.2418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2011 -0.2418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3899 -0.3932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3899 -0.3932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8677 0.2797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8677 0.2797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6429 -0.0034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6429 -0.0034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8542 0.5651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8542 0.5651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8899 -0.3785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8899 -0.3785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4473 -0.3092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4473 -0.3092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3118 -0.8950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3118 -0.8950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5837 -2.1931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5837 -2.1931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1825 0.1521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1825 0.1521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1693 -1.4735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1693 -1.4735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4374 0.8799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4374 0.8799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8493 1.5934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8493 1.5934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4374 2.3069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4374 2.3069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6732 1.5934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6732 1.5934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0852 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0852 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9090 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9090 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3215 1.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3215 1.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1465 1.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1465 1.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5590 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5590 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1465 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1465 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3215 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3215 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3118 -1.4989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3118 -1.4989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8527 -1.8112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8527 -1.8112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8346 -1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8346 -1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 13 17 1 0 0 0 0 | + | 13 17 1 0 0 0 0 |
| − | 18 19 1 1 0 0 0 | + | 18 19 1 1 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 18 25 1 0 0 0 0 | + | 18 25 1 0 0 0 0 |
| − | 19 26 1 0 0 0 0 | + | 19 26 1 0 0 0 0 |
| − | 20 27 1 0 0 0 0 | + | 20 27 1 0 0 0 0 |
| − | 3 28 1 0 0 0 0 | + | 3 28 1 0 0 0 0 |
| − | 1 29 1 0 0 0 0 | + | 1 29 1 0 0 0 0 |
| − | 2 30 1 0 0 0 0 | + | 2 30 1 0 0 0 0 |
| − | 29 21 1 0 0 0 0 | + | 29 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 31 24 1 0 0 0 0 | + | 31 24 1 0 0 0 0 |
| − | 27 42 1 0 0 0 0 | + | 27 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1C3AGS0003 | + | ID FL1C3AGS0003 |
| − | KNApSAcK_ID C00014501 | + | KNApSAcK_ID C00014501 |
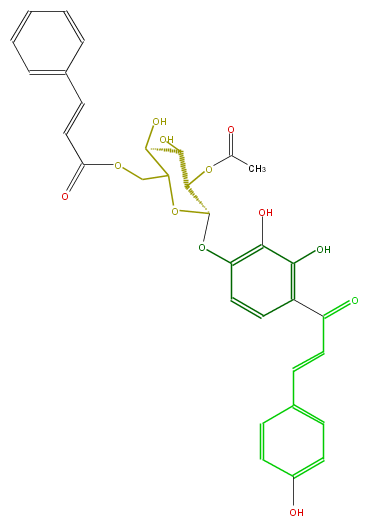
| − | NAME 4,2',3',4'-Tetrahydroxychalcone 4'-O-(2''-O-acetyl-6''-O-cinnamoyl)glucoside | + | NAME 4,2',3',4'-Tetrahydroxychalcone 4'-O-(2''-O-acetyl-6''-O-cinnamoyl)glucoside |
| − | CAS_RN 213473-58-6 | + | CAS_RN 213473-58-6 |
| − | FORMULA C32H30O12 | + | FORMULA C32H30O12 |
| − | EXACTMASS 606.173726424 | + | EXACTMASS 606.173726424 |
| − | AVERAGEMASS 606.5734 | + | AVERAGEMASS 606.5734 |
| − | SMILES C(c(c4)ccc(O)c4)=CC(=O)c(c3O)ccc(c(O)3)OC(O1)C(C(C(O)C1COC(=O)C=Cc(c2)cccc2)O)OC(C)=O | + | SMILES C(c(c4)ccc(O)c4)=CC(=O)c(c3O)ccc(c(O)3)OC(O1)C(C(C(O)C1COC(=O)C=Cc(c2)cccc2)O)OC(C)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 47 0 0 0 0 0 0 0 0999 V2000
0.8692 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8692 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5837 -1.4819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2981 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2981 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5837 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0126 -1.4819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7271 -1.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7271 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4416 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1560 -0.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8705 0.1681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8705 0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1560 1.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4416 0.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0126 -2.3069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5590 1.3906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4542 0.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9763 -0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2011 -0.2418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3899 -0.3932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8677 0.2797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6429 -0.0034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8542 0.5651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8899 -0.3785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4473 -0.3092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3118 -0.8950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5837 -2.1931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1825 0.1521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1693 -1.4735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4374 0.8799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8493 1.5934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4374 2.3069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6732 1.5934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0852 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9090 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3215 1.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1465 1.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.5590 0.8799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1465 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3215 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3118 -1.4989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8527 -1.8112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8346 -1.7744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
13 17 1 0 0 0 0
18 19 1 1 0 0 0
19 20 1 1 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 18 1 0 0 0 0
23 24 1 0 0 0 0
18 25 1 0 0 0 0
19 26 1 0 0 0 0
20 27 1 0 0 0 0
3 28 1 0 0 0 0
1 29 1 0 0 0 0
2 30 1 0 0 0 0
29 21 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
31 24 1 0 0 0 0
27 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
S SKP 8
ID FL1C3AGS0003
KNApSAcK_ID C00014501
NAME 4,2',3',4'-Tetrahydroxychalcone 4'-O-(2''-O-acetyl-6''-O-cinnamoyl)glucoside
CAS_RN 213473-58-6
FORMULA C32H30O12
EXACTMASS 606.173726424
AVERAGEMASS 606.5734
SMILES C(c(c4)ccc(O)c4)=CC(=O)c(c3O)ccc(c(O)3)OC(O1)C(C(C(O)C1COC(=O)C=Cc(c2)cccc2)O)OC(C)=O
M END