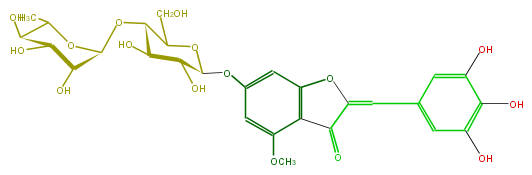
Mol:FL1ABGGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.9833 -0.9649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9833 -0.9649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9833 -0.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9833 -0.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2688 0.2725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2688 0.2725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4457 -0.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4457 -0.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4457 -0.9649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4457 -0.9649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2688 -1.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2688 -1.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7679 -1.2198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7679 -1.2198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2528 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2528 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7679 0.1150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7679 0.1150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9332 0.2519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9332 0.2519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8605 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8605 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0768 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0768 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4900 -1.2679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4900 -1.2679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3162 -1.2679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3162 -1.2679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7295 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7295 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3162 0.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3162 0.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4900 0.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4900 0.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5544 -0.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5544 -0.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9664 -1.9610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9664 -1.9610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7288 0.8777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7288 0.8777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0863 1.5421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0863 1.5421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9810 0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9810 0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2012 0.5829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2012 0.5829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5828 0.2346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5828 0.2346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7714 0.9391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7714 0.9391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4984 0.9668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4984 0.9668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7729 1.5820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7729 1.5820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5235 1.0041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5235 1.0041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7887 -0.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7887 -0.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9569 0.3557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9569 0.3557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2342 0.7730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2342 0.7730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0438 0.9756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0438 0.9756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6281 1.5779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6281 1.5779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.3509 1.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.3509 1.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5414 0.9580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5414 0.9580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5544 1.7073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5544 1.7073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0159 1.7130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0159 1.7130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.3369 0.8462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.3369 0.8462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3326 -0.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3326 -0.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7288 -1.9824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7288 -1.9824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2837 -2.0594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2837 -2.0594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2667 -2.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2667 -2.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7105 1.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7105 1.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5133 2.6269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5133 2.6269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 1 7 1 0 0 0 0 | + | 1 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 2 1 0 0 0 0 | + | 9 2 1 0 0 0 0 |
| − | 4 10 1 0 0 0 0 | + | 4 10 1 0 0 0 0 |
| − | 8 11 2 0 0 0 0 | + | 8 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 7 19 2 0 0 0 0 | + | 7 19 2 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 31 27 1 0 0 0 0 | + | 31 27 1 0 0 0 0 |
| − | 10 24 1 0 0 0 0 | + | 10 24 1 0 0 0 0 |
| − | 14 40 1 0 0 0 0 | + | 14 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 6 41 1 0 0 0 0 | + | 6 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 46 -0.0149 0.6820 | + | M SBV 1 46 -0.0149 0.6820 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 48 0.2121 -0.8574 | + | M SBV 2 48 0.2121 -0.8574 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL1ABGGS0001 | + | ID FL1ABGGS0001 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES CC(C(O)5)OC(C(C(O)5)O)OC(C4O)C(OC(C4O)Oc(c3)cc(c(c(OC)3)1)oc(=Cc(c2)cc(O)c(c(O)2)O)c(=O)1)CO | + | SMILES CC(C(O)5)OC(C(C(O)5)O)OC(C4O)C(OC(C4O)Oc(c3)cc(c(c(OC)3)1)oc(=Cc(c2)cc(O)c(c(O)2)O)c(=O)1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
0.9833 -0.9649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9833 -0.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2688 0.2725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4457 -0.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4457 -0.9649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2688 -1.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7679 -1.2198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2528 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7679 0.1150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9332 0.2519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8605 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0768 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4900 -1.2679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3162 -1.2679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7295 -0.5523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3162 0.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4900 0.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5544 -0.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9664 -1.9610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7288 0.8777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0863 1.5421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9810 0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2012 0.5829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5828 0.2346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7714 0.9391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4984 0.9668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7729 1.5820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5235 1.0041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7887 -0.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9569 0.3557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2342 0.7730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0438 0.9756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.6281 1.5779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.3509 1.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5414 0.9580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.5544 1.7073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.0159 1.7130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.3369 0.8462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3326 -0.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7288 -1.9824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2837 -2.0594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2667 -2.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7105 1.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5133 2.6269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
1 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 2 1 0 0 0 0
4 10 1 0 0 0 0
8 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
7 19 2 0 0 0 0
16 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
35 38 1 0 0 0 0
30 39 1 0 0 0 0
31 27 1 0 0 0 0
10 24 1 0 0 0 0
14 40 1 0 0 0 0
41 42 1 0 0 0 0
6 41 1 0 0 0 0
43 44 1 0 0 0 0
26 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 OCH3
M SBV 1 46 -0.0149 0.6820
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 ^CH2OH
M SBV 2 48 0.2121 -0.8574
S SKP 5
ID FL1ABGGS0001
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES CC(C(O)5)OC(C(C(O)5)O)OC(C4O)C(OC(C4O)Oc(c3)cc(c(c(OC)3)1)oc(=Cc(c2)cc(O)c(c(O)2)O)c(=O)1)CO
M END