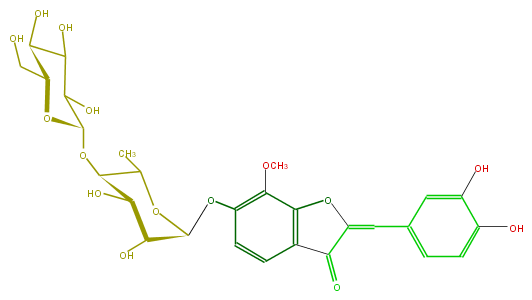
Mol:FL1A3CGS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -5.7626 1.9687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7626 1.9687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7737 -2.2638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7737 -2.2638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7737 -1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7737 -1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0600 -1.0283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0600 -1.0283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6533 -1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6533 -1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6533 -2.2638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6533 -2.2638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0600 -2.6757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0600 -2.6757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5570 -2.5184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5570 -2.5184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0414 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0414 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5570 -1.1856 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5570 -1.1856 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2457 -1.2075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2457 -1.2075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6482 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6482 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4708 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4708 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8835 -2.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8835 -2.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7086 -2.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7086 -2.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1211 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1211 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7086 -1.1374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7086 -1.1374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8835 -1.1374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8835 -1.1374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9450 -1.8519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9450 -1.8519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7554 -3.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7554 -3.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1204 -0.4239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1204 -0.4239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9118 -0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9118 -0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8914 -0.6369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8914 -0.6369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1155 -1.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1155 -1.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7576 -2.1618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7576 -2.1618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7778 -2.0588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7778 -2.0588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5535 -1.4519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5535 -1.4519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3560 -0.0895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3560 -0.0895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3222 -2.5047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3222 -2.5047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9126 -1.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9126 -1.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2708 -0.1161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2708 -0.1161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6861 2.1963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6861 2.1963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5606 2.4614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5606 2.4614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1165 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1165 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1412 0.7499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1412 0.7499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2670 0.4843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2670 0.4843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7109 1.2830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7109 1.2830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1394 0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1394 0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7748 2.9093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7748 2.9093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3471 3.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3471 3.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9450 2.6496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9450 2.6496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0600 -0.3596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0600 -0.3596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6359 0.8880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6359 0.8880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 2 0 0 0 0 | + | 6 7 2 0 0 0 0 |
| − | 7 2 1 0 0 0 0 | + | 7 2 1 0 0 0 0 |
| − | 2 8 1 0 0 0 0 | + | 2 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 5 11 1 0 0 0 0 | + | 5 11 1 0 0 0 0 |
| − | 9 12 2 0 0 0 0 | + | 9 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 8 20 2 0 0 0 0 | + | 8 20 2 0 0 0 0 |
| − | 17 21 1 0 0 0 0 | + | 17 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 26 11 1 0 0 0 0 | + | 26 11 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 34 1 1 0 0 0 0 | + | 34 1 1 0 0 0 0 |
| − | 1 41 1 0 0 0 0 | + | 1 41 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 4 42 1 0 0 0 0 | + | 4 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 0.0000 -0.6687 | + | M SBV 1 47 0.0000 -0.6687 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL1A3CGS0009 | + | ID FL1A3CGS0009 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES C(O)C(C(O)5)OC(C(O)C(O)5)OC(C(C)1)C(O)C(C(Oc(c(OC)2)ccc(c(=O)3)c(oc3=Cc(c4)ccc(O)c(O)4)2)O1)O | + | SMILES C(O)C(C(O)5)OC(C(O)C(O)5)OC(C(C)1)C(O)C(C(Oc(c(OC)2)ccc(c(=O)3)c(oc3=Cc(c4)ccc(O)c(O)4)2)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-5.7626 1.9687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7737 -2.2638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7737 -1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0600 -1.0283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6533 -1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6533 -2.2638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0600 -2.6757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5570 -2.5184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0414 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5570 -1.1856 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2457 -1.2075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6482 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4708 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8835 -2.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7086 -2.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1211 -1.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7086 -1.1374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8835 -1.1374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9450 -1.8519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7554 -3.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1204 -0.4239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9118 -0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8914 -0.6369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1155 -1.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7576 -2.1618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7778 -2.0588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5535 -1.4519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3560 -0.0895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3222 -2.5047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9126 -1.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2708 -0.1161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6861 2.1963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5606 2.4614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1165 1.6628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1412 0.7499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2670 0.4843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7109 1.2830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1394 0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7748 2.9093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3471 3.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9450 2.6496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0600 -0.3596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6359 0.8880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 2 1 0 0 0 0
2 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
5 11 1 0 0 0 0
9 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
16 19 1 0 0 0 0
8 20 2 0 0 0 0
17 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
26 11 1 0 0 0 0
22 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
23 31 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
31 36 1 0 0 0 0
37 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
34 1 1 0 0 0 0
1 41 1 0 0 0 0
42 43 1 0 0 0 0
4 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 0.0000 -0.6687
S SKP 5
ID FL1A3CGS0009
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES C(O)C(C(O)5)OC(C(O)C(O)5)OC(C(C)1)C(O)C(C(Oc(c(OC)2)ccc(c(=O)3)c(oc3=Cc(c4)ccc(O)c(O)4)2)O1)O
M END