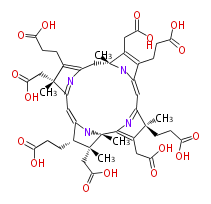
Mol:BMCCPPPC0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 64 68 0 0 1 0 0 0 0 0999 V2000 | + | 64 68 0 0 1 0 0 0 0 0999 V2000 |
| − | -0.2046 2.0961 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2046 2.0961 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2941 2.9599 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.2941 2.9599 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.2698 2.7525 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2698 2.7525 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.3741 1.7606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3741 1.7606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9261 1.1474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9261 1.1474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1811 0.3628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1811 0.3628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9881 -0.2235 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 2.9881 -0.2235 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.6798 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6798 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6823 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6823 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0149 -1.6571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0149 -1.6571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2079 -1.8286 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.2079 -1.8286 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.4595 -2.5699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4595 -2.5699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3707 -2.1641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3707 -2.1641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2665 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2665 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6790 -0.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6790 -0.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7653 0.3628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7653 0.3628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4327 1.1041 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4327 1.1041 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9340 1.9679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9340 1.9679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9582 1.7606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9582 1.7606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6204 2.0961 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6204 2.0961 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0948 -0.4577 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0948 -0.4577 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5990 -1.6571 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5990 -1.6571 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5103 1.1474 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5103 1.1474 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0115 2.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0115 2.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4203 3.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4203 3.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4657 3.7669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4657 3.7669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1474 4.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1474 4.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8829 3.3045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8829 3.3045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6676 3.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6676 3.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2806 3.6017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2806 3.6017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4729 0.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4729 0.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7726 -0.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7726 -0.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3858 0.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3858 0.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1647 -1.8396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1647 -1.8396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9852 -1.7534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9852 -1.7534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4701 -2.4208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4701 -2.4208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2880 -3.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2880 -3.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9011 -3.9289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9011 -3.9289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9283 -3.0634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9283 -3.0634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9283 -3.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9283 -3.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6428 -4.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6428 -4.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0457 0.5520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0457 0.5520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1471 1.5166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1471 1.5166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8616 1.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8616 1.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5761 1.5166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5761 1.5166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2695 2.7216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2695 2.7216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0900 2.8078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0900 2.8078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9616 -2.1641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9616 -2.1641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3572 -3.8884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3572 -3.8884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6428 -5.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6428 -5.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1346 -3.1744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1346 -3.1744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2906 -2.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2906 -2.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2142 0.8806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2142 0.8806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1704 -0.1813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1704 -0.1813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1091 4.4086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1091 4.4086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0653 3.3467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0653 3.3467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6857 -3.6739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6857 -3.6739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7295 -4.7358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7295 -4.7358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5761 2.3416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5761 2.3416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2906 1.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2906 1.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5749 2.1404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5749 2.1404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4256 3.5615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4256 3.5615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9320 4.0639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9320 4.0639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0241 5.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0241 5.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17 43 1 0 0 0 0 | + | 17 43 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 36 51 1 0 0 0 0 | + | 36 51 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 36 52 2 0 0 0 0 | + | 36 52 2 0 0 0 0 |
| − | 22 11 1 0 0 0 0 | + | 22 11 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 33 53 1 0 0 0 0 | + | 33 53 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 33 54 2 0 0 0 0 | + | 33 54 2 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 16 2 0 0 0 0 | + | 23 16 2 0 0 0 0 |
| − | 30 55 1 0 0 0 0 | + | 30 55 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 30 56 2 0 0 0 0 | + | 30 56 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 19 23 1 0 0 0 0 | + | 19 23 1 0 0 0 0 |
| − | 16 15 1 0 0 0 0 | + | 16 15 1 0 0 0 0 |
| − | 15 14 2 0 0 0 0 | + | 15 14 2 0 0 0 0 |
| − | 12 37 1 0 0 0 0 | + | 12 37 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 11 10 1 0 0 0 0 | + | 11 10 1 0 0 0 0 |
| − | 38 57 1 0 0 0 0 | + | 38 57 1 0 0 0 0 |
| − | 10 9 1 0 0 0 0 | + | 10 9 1 0 0 0 0 |
| − | 38 58 2 0 0 0 0 | + | 38 58 2 0 0 0 0 |
| − | 6 21 2 0 0 0 0 | + | 6 21 2 0 0 0 0 |
| − | 7 31 1 4 0 0 0 | + | 7 31 1 4 0 0 0 |
| − | 3 28 1 4 0 0 0 | + | 3 28 1 4 0 0 0 |
| − | 45 59 1 0 0 0 0 | + | 45 59 1 0 0 0 0 |
| − | 9 8 2 0 0 0 0 | + | 9 8 2 0 0 0 0 |
| − | 45 60 2 0 0 0 0 | + | 45 60 2 0 0 0 0 |
| − | 8 7 1 0 0 0 0 | + | 8 7 1 0 0 0 0 |
| − | 18 46 1 0 0 0 0 | + | 18 46 1 0 0 0 0 |
| − | 7 6 1 0 0 0 0 | + | 7 6 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 21 9 1 0 0 0 0 | + | 21 9 1 0 0 0 0 |
| − | 47 61 1 0 0 0 0 | + | 47 61 1 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 47 62 2 0 0 0 0 | + | 47 62 2 0 0 0 0 |
| − | 13 39 1 0 0 0 0 | + | 13 39 1 0 0 0 0 |
| − | 20 4 1 0 0 0 0 | + | 20 4 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 17 42 1 4 0 0 0 | + | 17 42 1 4 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 7 32 1 0 0 0 0 | + | 7 32 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 8 34 1 0 0 0 0 | + | 8 34 1 0 0 0 0 |
| − | 2 26 1 0 0 0 0 | + | 2 26 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 2 25 1 4 0 0 0 | + | 2 25 1 4 0 0 0 |
| − | 41 49 2 0 0 0 0 | + | 41 49 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 27 63 1 0 0 0 0 | + | 27 63 1 0 0 0 0 |
| − | 41 50 1 0 0 0 0 | + | 41 50 1 0 0 0 0 |
| − | 27 64 2 0 0 0 0 | + | 27 64 2 0 0 0 0 |
| − | 11 48 1 4 0 0 0 | + | 11 48 1 4 0 0 0 |
| − | 1 24 1 4 0 0 0 | + | 1 24 1 4 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPPPC0006 | + | ID BMCCPPPC0006 |
| − | NAME Precorrin 6X | + | NAME Precorrin 6X |
| − | FORMULA C44H54N4O16 | + | FORMULA C44H54N4O16 |
| − | EXACTMASS 894.3534 | + | EXACTMASS 894.3534 |
| − | AVERAGEMASS 894.9169 | + | AVERAGEMASS 894.9169 |
| − | SMILES C(=N3)([C@@](CC(O)=O)(C)4)C=C([C@H]5CCC(O)=O)N[C@]([C@@]5(CC(O)=O)C)(C)C(N=1)=C(CC(O)=O)[C@@](C1C=C(C(CCC(O)=O)=2)N[C@](C)(CC3=C4CCC(O)=O)C2CC(O)=O)(CCC(O)=O)C | + | SMILES C(=N3)([C@@](CC(O)=O)(C)4)C=C([C@H]5CCC(O)=O)N[C@]([C@@]5(CC(O)=O)C)(C)C(N=1)=C(CC(O)=O)[C@@](C1C=C(C(CCC(O)=O)=2)N[C@](C)(CC3=C4CCC(O)=O)C2CC(O)=O)(CCC(O)=O)C |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06320 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06320 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
64 68 0 0 1 0 0 0 0 0999 V2000
-0.2046 2.0961 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2941 2.9599 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.2698 2.7525 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.3741 1.7606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9261 1.1474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1811 0.3628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9881 -0.2235 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.6798 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6823 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0149 -1.6571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2079 -1.8286 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.4595 -2.5699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3707 -2.1641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2665 -1.1722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6790 -0.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7653 0.3628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4327 1.1041 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9340 1.9679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9582 1.7606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6204 2.0961 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.0948 -0.4577 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.5990 -1.6571 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.5103 1.1474 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.0115 2.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4203 3.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4657 3.7669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1474 4.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8829 3.3045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6676 3.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2806 3.6017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4729 0.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7726 -0.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3858 0.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1647 -1.8396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9852 -1.7534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4701 -2.4208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2880 -3.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9011 -3.9289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9283 -3.0634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9283 -3.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6428 -4.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0457 0.5520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1471 1.5166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8616 1.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5761 1.5166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2695 2.7216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0900 2.8078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9616 -2.1641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3572 -3.8884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6428 -5.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1346 -3.1744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2906 -2.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2142 0.8806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1704 -0.1813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1091 4.4086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0653 3.3467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6857 -3.6739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7295 -4.7358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5761 2.3416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2906 1.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5749 2.1404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4256 3.5615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9320 4.0639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0241 5.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17 43 1 0 0 0 0
11 12 1 0 0 0 0
35 36 1 0 0 0 0
12 13 2 0 0 0 0
36 51 1 0 0 0 0
13 14 1 0 0 0 0
36 52 2 0 0 0 0
22 11 1 0 0 0 0
32 33 1 0 0 0 0
18 19 2 0 0 0 0
33 53 1 0 0 0 0
28 29 1 0 0 0 0
33 54 2 0 0 0 0
43 44 1 0 0 0 0
29 30 1 0 0 0 0
23 16 2 0 0 0 0
30 55 1 0 0 0 0
16 17 1 0 0 0 0
30 56 2 0 0 0 0
17 18 1 0 0 0 0
19 23 1 0 0 0 0
16 15 1 0 0 0 0
15 14 2 0 0 0 0
12 37 1 0 0 0 0
44 45 1 0 0 0 0
37 38 1 0 0 0 0
11 10 1 0 0 0 0
38 57 1 0 0 0 0
10 9 1 0 0 0 0
38 58 2 0 0 0 0
6 21 2 0 0 0 0
7 31 1 4 0 0 0
3 28 1 4 0 0 0
45 59 1 0 0 0 0
9 8 2 0 0 0 0
45 60 2 0 0 0 0
8 7 1 0 0 0 0
18 46 1 0 0 0 0
7 6 1 0 0 0 0
46 47 1 0 0 0 0
21 9 1 0 0 0 0
47 61 1 0 0 0 0
2 1 1 0 0 0 0
47 62 2 0 0 0 0
13 39 1 0 0 0 0
20 4 1 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
1 20 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
17 42 1 4 0 0 0
1 19 1 0 0 0 0
39 40 1 0 0 0 0
7 32 1 0 0 0 0
40 41 1 0 0 0 0
8 34 1 0 0 0 0
2 26 1 0 0 0 0
34 35 1 0 0 0 0
2 25 1 4 0 0 0
41 49 2 0 0 0 0
26 27 1 0 0 0 0
14 22 1 0 0 0 0
27 63 1 0 0 0 0
41 50 1 0 0 0 0
27 64 2 0 0 0 0
11 48 1 4 0 0 0
1 24 1 4 0 0 0
S SKP 7
ID BMCCPPPC0006
NAME Precorrin 6X
FORMULA C44H54N4O16
EXACTMASS 894.3534
AVERAGEMASS 894.9169
SMILES C(=N3)([C@@](CC(O)=O)(C)4)C=C([C@H]5CCC(O)=O)N[C@]([C@@]5(CC(O)=O)C)(C)C(N=1)=C(CC(O)=O)[C@@](C1C=C(C(CCC(O)=O)=2)N[C@](C)(CC3=C4CCC(O)=O)C2CC(O)=O)(CCC(O)=O)C
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06320
M END