Mol:BMCCPPPC0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 62 66 0 0 1 0 0 0 0 0999 V2000 | + | 62 66 0 0 1 0 0 0 0 0999 V2000 |
| − | 7.1714 2.5470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1714 2.5470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0913 3.0974 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 6.0913 3.0974 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 5.2340 2.2402 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 5.2340 2.2402 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.7844 1.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7844 1.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5893 0.1792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5893 0.1792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7844 -0.8016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7844 -0.8016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2340 -1.8818 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 5.2340 -1.8818 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 6.0913 -2.7390 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 6.0913 -2.7390 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 7.1714 -2.1886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1714 -2.1886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.1522 -2.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.1522 -2.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1330 -2.1886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1330 -2.1886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2132 -2.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2132 -2.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.0704 -1.8818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.0704 -1.8818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5200 -0.8016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5200 -0.8016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.7151 0.1792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.7151 0.1792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5200 1.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5200 1.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.0704 2.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.0704 2.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2132 3.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2132 3.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1330 2.5470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1330 2.5470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.1522 2.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.1522 2.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3400 1.9914 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3400 1.9914 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3400 -1.6331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3400 -1.6331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9645 -1.6331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9645 -1.6331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9645 1.9914 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9645 1.9914 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5452 3.9884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5452 3.9884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3841 3.8045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3841 3.8045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6430 4.7704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6430 4.7704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2463 2.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2463 2.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6170 1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6170 1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6293 1.7759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6293 1.7759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3430 -1.4278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3430 -1.4278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5269 -2.5889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5269 -2.5889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5610 -2.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5610 -2.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9348 -3.7267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9348 -3.7267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0012 -4.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0012 -4.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8448 -5.0728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8448 -5.0728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.3696 -3.7267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.3696 -3.7267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.5925 -4.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.5925 -4.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.0581 -2.0382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.0581 -2.0382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4165 -2.9718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4165 -2.9718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.4042 -3.1282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.4042 -3.1282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.0581 2.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.0581 2.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4165 3.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4165 3.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.4042 3.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.4042 3.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.3696 4.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.3696 4.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.5925 4.7144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.5925 4.7144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.0335 -2.3511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.0335 -2.3511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.7625 -4.0618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.7625 -4.0618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6220 -5.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6220 -5.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9112 -5.4311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9112 -5.4311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3022 -1.3641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3022 -1.3641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8539 -3.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8539 -3.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 0.9987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 0.9987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2710 2.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2710 2.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6089 5.0292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6089 5.0292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9359 5.4775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9359 5.4775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.6589 -3.9977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.6589 -3.9977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.7489 -5.3437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.7489 -5.3437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.0335 2.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.0335 2.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.7625 4.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.7625 4.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.6589 4.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.6589 4.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.7489 5.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.7489 5.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21 4 1 0 0 0 0 | + | 21 4 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 1 20 2 0 0 0 0 | + | 1 20 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 7 32 1 6 0 0 0 | + | 7 32 1 6 0 0 0 |
| − | 2 26 1 6 0 0 0 | + | 2 26 1 6 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 8 34 1 4 0 0 0 | + | 8 34 1 4 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 41 47 2 0 0 0 0 | + | 41 47 2 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 41 48 1 0 0 0 0 | + | 41 48 1 0 0 0 0 |
| − | 17 42 1 0 0 0 0 | + | 17 42 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 36 49 1 0 0 0 0 | + | 36 49 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 36 50 2 0 0 0 0 | + | 36 50 2 0 0 0 0 |
| − | 23 11 1 0 0 0 0 | + | 23 11 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 33 51 1 0 0 0 0 | + | 33 51 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 33 52 2 0 0 0 0 | + | 33 52 2 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 24 16 1 0 0 0 0 | + | 24 16 1 0 0 0 0 |
| − | 30 53 1 0 0 0 0 | + | 30 53 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 30 54 2 0 0 0 0 | + | 30 54 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 19 24 1 0 0 0 0 | + | 19 24 1 0 0 0 0 |
| − | 27 55 1 0 0 0 0 | + | 27 55 1 0 0 0 0 |
| − | 16 15 1 0 0 0 0 | + | 16 15 1 0 0 0 0 |
| − | 27 56 2 0 0 0 0 | + | 27 56 2 0 0 0 0 |
| − | 15 14 1 0 0 0 0 | + | 15 14 1 0 0 0 0 |
| − | 12 37 1 0 0 0 0 | + | 12 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 11 10 1 0 0 0 0 | + | 11 10 1 0 0 0 0 |
| − | 38 57 1 0 0 0 0 | + | 38 57 1 0 0 0 0 |
| − | 10 9 2 0 0 0 0 | + | 10 9 2 0 0 0 0 |
| − | 38 58 2 0 0 0 0 | + | 38 58 2 0 0 0 0 |
| − | 6 22 2 0 0 0 0 | + | 6 22 2 0 0 0 0 |
| − | 7 31 1 1 0 0 0 | + | 7 31 1 1 0 0 0 |
| − | 20 19 1 0 0 0 0 | + | 20 19 1 0 0 0 0 |
| − | 3 28 1 4 0 0 0 | + | 3 28 1 4 0 0 0 |
| − | 9 8 1 0 0 0 0 | + | 9 8 1 0 0 0 0 |
| − | 8 7 1 0 0 0 0 | + | 8 7 1 0 0 0 0 |
| − | 18 45 1 0 0 0 0 | + | 18 45 1 0 0 0 0 |
| − | 7 6 1 0 0 0 0 | + | 7 6 1 0 0 0 0 |
| − | 22 9 1 0 0 0 0 | + | 22 9 1 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 13 39 1 0 0 0 0 | + | 13 39 1 0 0 0 0 |
| − | 2 25 1 1 0 0 0 | + | 2 25 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 59 1 0 0 0 0 | + | 44 59 1 0 0 0 0 |
| − | 44 60 2 0 0 0 0 | + | 44 60 2 0 0 0 0 |
| − | 46 61 1 0 0 0 0 | + | 46 61 1 0 0 0 0 |
| − | 46 62 2 0 0 0 0 | + | 46 62 2 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPPPC0001 | + | ID BMCCPPPC0001 |
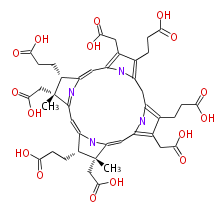
| − | NAME Precorrin 2 | + | NAME Precorrin 2 |
| − | FORMULA C42H48N4O16 | + | FORMULA C42H48N4O16 |
| − | EXACTMASS 864.3065 | + | EXACTMASS 864.3065 |
| − | AVERAGEMASS 864.8478 | + | AVERAGEMASS 864.8478 |
| − | SMILES c(c35)(CC(O)=O)c(c(n5)Cc(c4CCC(O)=O)nc(c4CC(O)=O)C=C(N1)[C@@]([C@@H](C(=CC([C@@](CC(O)=O)(C)2)=NC(=C3)[C@H]2CCC(O)=O)1)CCC(O)=O)(CC(O)=O)C)CCC(O)=O | + | SMILES c(c35)(CC(O)=O)c(c(n5)Cc(c4CCC(O)=O)nc(c4CC(O)=O)C=C(N1)[C@@]([C@@H](C(=CC([C@@](CC(O)=O)(C)2)=NC(=C3)[C@H]2CCC(O)=O)1)CCC(O)=O)(CC(O)=O)C)CCC(O)=O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C02463 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C02463 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
62 66 0 0 1 0 0 0 0 0999 V2000
7.1714 2.5470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0913 3.0974 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
5.2340 2.2402 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.7844 1.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5893 0.1792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7844 -0.8016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2340 -1.8818 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
6.0913 -2.7390 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
7.1714 -2.1886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.1522 -2.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1330 -2.1886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2132 -2.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.0704 -1.8818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.5200 -0.8016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.7151 0.1792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.5200 1.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.0704 2.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2132 3.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1330 2.5470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.1522 2.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3400 1.9914 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
6.3400 -1.6331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.9645 -1.6331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.9645 1.9914 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
6.5452 3.9884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3841 3.8045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6430 4.7704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2463 2.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6170 1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6293 1.7759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3430 -1.4278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5269 -2.5889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5610 -2.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9348 -3.7267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0012 -4.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8448 -5.0728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3696 -3.7267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5925 -4.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.0581 -2.0382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.4165 -2.9718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.4042 -3.1282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.0581 2.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.4165 3.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.4042 3.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3696 4.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5925 4.7144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.0335 -2.3511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.7625 -4.0618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6220 -5.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9112 -5.4311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3022 -1.3641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8539 -3.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 0.9987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2710 2.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.6089 5.0292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9359 5.4775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.6589 -3.9977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.7489 -5.3437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.0335 2.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.7625 4.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.6589 4.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.7489 5.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21 4 1 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
1 21 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
1 20 2 0 0 0 0
39 40 1 0 0 0 0
7 32 1 6 0 0 0
2 26 1 6 0 0 0
40 41 1 0 0 0 0
8 34 1 4 0 0 0
34 35 1 0 0 0 0
41 47 2 0 0 0 0
14 23 1 0 0 0 0
41 48 1 0 0 0 0
17 42 1 0 0 0 0
11 12 2 0 0 0 0
35 36 1 0 0 0 0
12 13 1 0 0 0 0
36 49 1 0 0 0 0
13 14 2 0 0 0 0
36 50 2 0 0 0 0
23 11 1 0 0 0 0
32 33 1 0 0 0 0
18 19 2 0 0 0 0
33 51 1 0 0 0 0
28 29 1 0 0 0 0
33 52 2 0 0 0 0
29 30 1 0 0 0 0
24 16 1 0 0 0 0
30 53 1 0 0 0 0
16 17 2 0 0 0 0
30 54 2 0 0 0 0
17 18 1 0 0 0 0
26 27 1 0 0 0 0
19 24 1 0 0 0 0
27 55 1 0 0 0 0
16 15 1 0 0 0 0
27 56 2 0 0 0 0
15 14 1 0 0 0 0
12 37 1 0 0 0 0
37 38 1 0 0 0 0
11 10 1 0 0 0 0
38 57 1 0 0 0 0
10 9 2 0 0 0 0
38 58 2 0 0 0 0
6 22 2 0 0 0 0
7 31 1 1 0 0 0
20 19 1 0 0 0 0
3 28 1 4 0 0 0
9 8 1 0 0 0 0
8 7 1 0 0 0 0
18 45 1 0 0 0 0
7 6 1 0 0 0 0
22 9 1 0 0 0 0
2 1 1 0 0 0 0
13 39 1 0 0 0 0
2 25 1 1 0 0 0
45 46 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 59 1 0 0 0 0
44 60 2 0 0 0 0
46 61 1 0 0 0 0
46 62 2 0 0 0 0
S SKP 7
ID BMCCPPPC0001
NAME Precorrin 2
FORMULA C42H48N4O16
EXACTMASS 864.3065
AVERAGEMASS 864.8478
SMILES c(c35)(CC(O)=O)c(c(n5)Cc(c4CCC(O)=O)nc(c4CC(O)=O)C=C(N1)[C@@]([C@@H](C(=CC([C@@](CC(O)=O)(C)2)=NC(=C3)[C@H]2CCC(O)=O)1)CCC(O)=O)(CC(O)=O)C)CCC(O)=O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C02463
M END