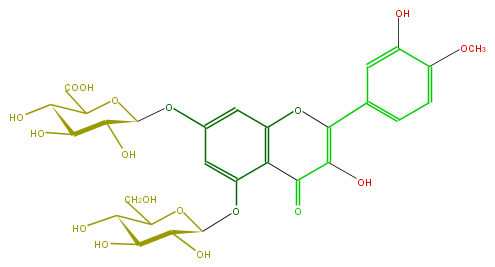
Mol:FL5FAEGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1022 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1022 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1022 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1022 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3876 -1.1024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3876 -1.1024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3269 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3269 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3269 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3269 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3876 0.5478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3876 0.5478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0415 -1.1024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0415 -1.1024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7560 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7560 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7560 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7560 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0415 0.5478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0415 0.5478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0428 -1.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0428 -1.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6557 0.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6557 0.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3839 0.2861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3839 0.2861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1122 0.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1122 0.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1122 1.5475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1122 1.5475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3839 1.9680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3839 1.9680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6557 1.5475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6557 1.5475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3883 -1.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3883 -1.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9557 0.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9557 0.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3839 2.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3839 2.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5011 -1.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5011 -1.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6949 0.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6949 0.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1704 -0.0416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1704 -0.0416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4152 0.2522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4152 0.2522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6866 0.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6866 0.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2160 0.7896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2160 0.7896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8768 0.4409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8768 0.4409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4046 0.3924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4046 0.3924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9322 0.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9322 0.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9826 -0.4742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9826 -0.4742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1720 -1.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1720 -1.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6475 -2.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6475 -2.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8923 -2.3078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8923 -2.3078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1636 -2.2999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1636 -2.2999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6931 -1.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6931 -1.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3538 -2.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3538 -2.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9595 -2.1957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9595 -2.1957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4381 -2.5776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4381 -2.5776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2313 -2.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2313 -2.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0215 -1.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0215 -1.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1827 -1.6208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1827 -1.6208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4370 1.0836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4370 1.0836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8093 1.4193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8093 1.4193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4601 2.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4601 2.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8916 1.9975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8916 1.9975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8093 1.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8093 1.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 27 42 1 0 0 0 0 | + | 27 42 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 15 45 1 0 0 0 0 | + | 15 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.6677 -0.5997 | + | M SBV 1 45 0.6677 -0.5997 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 42 43 44 | + | M SAL 2 3 42 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 48 0.5602 -0.6427 | + | M SBV 2 48 0.5602 -0.6427 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 45 46 | + | M SAL 3 2 45 46 |
| − | M SBL 3 1 50 | + | M SBL 3 1 50 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 50 -0.7794 -0.4501 | + | M SBV 3 50 -0.7794 -0.4501 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAEGS0003 | + | ID FL5FAEGS0003 |
| − | FORMULA C28H30O18 | + | FORMULA C28H30O18 |
| − | EXACTMASS 654.143214156 | + | EXACTMASS 654.143214156 |
| − | AVERAGEMASS 654.527 | + | AVERAGEMASS 654.527 |
| − | SMILES COc(c5)c(cc(c5)C(=C4O)Oc(c1)c(C4=O)c(OC(O3)C(C(O)C(O)C3CO)O)cc1OC(C(O)2)OC(C(O)=O)C(O)C2O)O | + | SMILES COc(c5)c(cc(c5)C(=C4O)Oc(c1)c(C4=O)c(OC(O3)C(C(O)C(O)C3CO)O)cc1OC(C(O)2)OC(C(O)=O)C(O)C2O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.1022 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1022 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3876 -1.1024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3269 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3269 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3876 0.5478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0415 -1.1024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7560 -0.6898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7560 0.1352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0415 0.5478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0428 -1.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6557 0.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3839 0.2861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1122 0.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1122 1.5475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3839 1.9680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6557 1.5475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3883 -1.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9557 0.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3839 2.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5011 -1.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6949 0.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1704 -0.0416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4152 0.2522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6866 0.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2160 0.7896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8768 0.4409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4046 0.3924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9322 0.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9826 -0.4742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1720 -1.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6475 -2.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8923 -2.3078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1636 -2.2999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6931 -1.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3538 -2.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9595 -2.1957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4381 -2.5776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2313 -2.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0215 -1.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1827 -1.6208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4370 1.0836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8093 1.4193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4601 2.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8916 1.9975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8093 1.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
4 3 1 0 0 0 0
1 19 1 0 0 0 0
16 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 18 1 0 0 0 0
40 41 1 0 0 0 0
36 40 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
27 42 1 0 0 0 0
45 46 1 0 0 0 0
15 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.6677 -0.5997
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 42 43 44
M SBL 2 1 48
M SMT 2 ^ COOH
M SBV 2 48 0.5602 -0.6427
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 45 46
M SBL 3 1 50
M SMT 3 OCH3
M SBV 3 50 -0.7794 -0.4501
S SKP 5
ID FL5FAEGS0003
FORMULA C28H30O18
EXACTMASS 654.143214156
AVERAGEMASS 654.527
SMILES COc(c5)c(cc(c5)C(=C4O)Oc(c1)c(C4=O)c(OC(O3)C(C(O)C(O)C3CO)O)cc1OC(C(O)2)OC(C(O)=O)C(O)C2O)O
M END