Mol:FL5FAAGS0036
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.1740 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1740 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1740 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1740 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8885 -2.1039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8885 -2.1039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6029 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6029 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6029 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6029 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8885 -0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8885 -0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3174 -2.1039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3174 -2.1039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0318 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0318 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0318 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0318 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3174 -0.4540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3174 -0.4540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3174 -2.7472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3174 -2.7472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7460 -0.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7460 -0.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4743 -0.8746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4743 -0.8746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2025 -0.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2025 -0.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2025 0.3867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2025 0.3867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4743 0.8071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4743 0.8071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7460 0.3867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7460 0.3867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8885 -2.9286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8885 -2.9286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5402 -0.4541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5402 -0.4541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9304 0.8070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9304 0.8070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8908 -2.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8908 -2.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9981 0.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9981 0.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7810 0.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7810 0.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2183 -0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2183 -0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0259 -0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0259 -0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2429 -0.8600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2429 -0.8600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8057 -0.3155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8057 -0.3155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3750 0.9313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3750 0.9313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9399 1.0243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9399 1.0243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9926 -0.0098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9926 -0.0098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6172 -1.0678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6172 -1.0678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7316 1.1705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7316 1.1705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1138 0.5684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1138 0.5684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3990 0.8734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3990 0.8734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6202 0.8237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6202 0.8237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2388 1.3056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2388 1.3056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9162 0.9989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9162 0.9989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5488 1.5145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5488 1.5145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9152 0.6483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9152 0.6483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7195 0.4115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7195 0.4115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1789 1.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1789 1.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9304 2.9286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9304 2.9286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 26 19 1 0 0 0 0 | + | 26 19 1 0 0 0 0 |
| − | 35 29 1 0 0 0 0 | + | 35 29 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 46 0.2627 -0.8881 | + | M SBV 1 46 0.2627 -0.8881 |
| − | S SKP 5 | + | S SKP 5 |
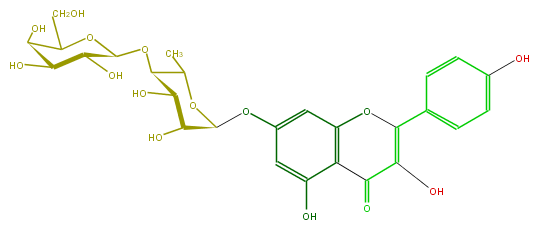
| − | ID FL5FAAGS0036 | + | ID FL5FAAGS0036 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES Oc(c5)ccc(c5)C(=C(O)4)Oc(c3C4=O)cc(cc3O)OC(O1)C(C(O)C(OC(O2)C(O)C(O)C(C2CO)O)C1C)O | + | SMILES Oc(c5)ccc(c5)C(=C(O)4)Oc(c3C4=O)cc(cc3O)OC(O1)C(C(O)C(OC(O2)C(O)C(O)C(C2CO)O)C1C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.1740 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1740 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8885 -2.1039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6029 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6029 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8885 -0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3174 -2.1039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0318 -1.6914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0318 -0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3174 -0.4540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3174 -2.7472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7460 -0.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4743 -0.8746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2025 -0.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2025 0.3867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4743 0.8071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7460 0.3867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8885 -2.9286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5402 -0.4541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9304 0.8070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8908 -2.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9981 0.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7810 0.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2183 -0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0259 -0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2429 -0.8600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8057 -0.3155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3750 0.9313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9399 1.0243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9926 -0.0098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6172 -1.0678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.7316 1.1705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1138 0.5684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3990 0.8734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6202 0.8237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2388 1.3056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9162 0.9989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5488 1.5145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9152 0.6483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7195 0.4115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1789 1.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9304 2.9286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
26 19 1 0 0 0 0
35 29 1 0 0 0 0
41 42 1 0 0 0 0
37 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^ CH2OH
M SBV 1 46 0.2627 -0.8881
S SKP 5
ID FL5FAAGS0036
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES Oc(c5)ccc(c5)C(=C(O)4)Oc(c3C4=O)cc(cc3O)OC(O1)C(C(O)C(OC(O2)C(O)C(O)C(C2CO)O)C1C)O
M END