Mol:FL3FAAGS0043
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 39 0 0 0 0 0 0 0 0999 V2000 | + | 36 39 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.6069 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6069 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6069 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6069 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1632 -0.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1632 -0.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7195 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7195 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7195 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7195 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1632 0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1632 0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2758 -0.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2758 -0.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8321 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8321 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8321 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8321 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2758 0.4020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2758 0.4020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2758 -1.3836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2758 -1.3836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3882 0.4019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3882 0.4019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9551 0.0745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9551 0.0745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5221 0.4019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5221 0.4019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5221 1.0566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5221 1.0566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9551 1.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9551 1.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3882 1.0566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3882 1.0566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0508 0.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0508 0.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0889 1.3838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0889 1.3838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1632 -1.3839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1632 -1.3839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7210 -0.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7210 -0.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2005 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2005 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7659 -0.6753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7659 -0.6753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1399 -0.4319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1399 -0.4319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4535 -0.5491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4535 -0.5491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9749 0.0136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9749 0.0136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6142 -0.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6142 -0.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0889 -0.4348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0889 -0.4348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4435 -0.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4435 -0.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7813 -1.0339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7813 -1.0339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7210 -0.6019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7210 -0.6019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4026 0.2982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4026 0.2982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4026 0.9282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4026 0.9282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9093 0.0057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9093 0.0057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6507 0.1825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6507 0.1825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4476 0.3960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4476 0.3960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 20 3 1 0 0 0 0 | + | 20 3 1 0 0 0 0 |
| − | 21 18 1 0 0 0 0 | + | 21 18 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 21 31 2 0 0 0 0 | + | 21 31 2 0 0 0 0 |
| − | 21 32 1 0 0 0 0 | + | 21 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 25 1 0 0 0 0 | + | 34 25 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 35 36 | + | M SAL 1 2 35 36 |
| − | M SBL 1 1 38 | + | M SBL 1 1 38 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 38 -8.0055 5.0580 | + | M SBV 1 38 -8.0055 5.0580 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0043 | + | ID FL3FAAGS0043 |
| − | KNApSAcK_ID C00004181 | + | KNApSAcK_ID C00004181 |
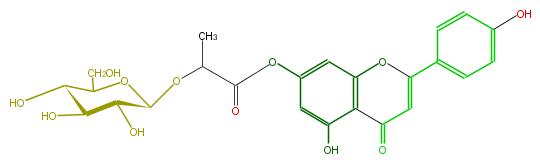
| − | NAME Apigenin 7-(2''-glucosyllactate) | + | NAME Apigenin 7-(2''-glucosyllactate) |
| − | CAS_RN 126394-94-3 | + | CAS_RN 126394-94-3 |
| − | FORMULA C24H24O12 | + | FORMULA C24H24O12 |
| − | EXACTMASS 504.126776232 | + | EXACTMASS 504.126776232 |
| − | AVERAGEMASS 504.44016 | + | AVERAGEMASS 504.44016 |
| − | SMILES C(=O)(C=1)c(c3O)c(cc(OC(C(C)OC(C(O)4)OC(C(O)C(O)4)CO)=O)c3)OC1c(c2)ccc(c2)O | + | SMILES C(=O)(C=1)c(c3O)c(cc(OC(C(C)OC(C(O)4)OC(C(O)C(O)4)CO)=O)c3)OC1c(c2)ccc(c2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 39 0 0 0 0 0 0 0 0999 V2000
0.6069 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6069 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1632 -0.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7195 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7195 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1632 0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2758 -0.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8321 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8321 0.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2758 0.4020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2758 -1.3836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3882 0.4019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9551 0.0745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5221 0.4019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5221 1.0566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9551 1.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3882 1.0566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0508 0.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0889 1.3838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1632 -1.3839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7210 -0.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2005 -0.1015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7659 -0.6753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1399 -0.4319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4535 -0.5491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9749 0.0136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6142 -0.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0889 -0.4348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4435 -0.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7813 -1.0339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7210 -0.6019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4026 0.2982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4026 0.9282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9093 0.0057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6507 0.1825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4476 0.3960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
20 3 1 0 0 0 0
21 18 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
21 31 2 0 0 0 0
21 32 1 0 0 0 0
32 33 1 0 0 0 0
32 34 1 0 0 0 0
34 25 1 0 0 0 0
27 35 1 0 0 0 0
35 36 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 35 36
M SBL 1 1 38
M SMT 1 ^CH2OH
M SBV 1 38 -8.0055 5.0580
S SKP 8
ID FL3FAAGS0043
KNApSAcK_ID C00004181
NAME Apigenin 7-(2''-glucosyllactate)
CAS_RN 126394-94-3
FORMULA C24H24O12
EXACTMASS 504.126776232
AVERAGEMASS 504.44016
SMILES C(=O)(C=1)c(c3O)c(cc(OC(C(C)OC(C(O)4)OC(C(O)C(O)4)CO)=O)c3)OC1c(c2)ccc(c2)O
M END